
| Sequence ID | X_DroMel_CAF1 |
|---|---|
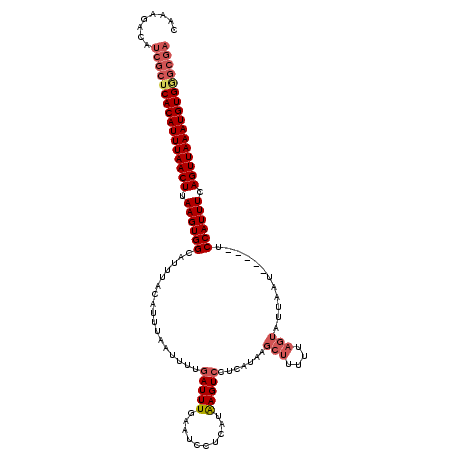
| Location | 15,927,424 – 15,927,539 |
| Length | 115 |
| Max. P | 0.999176 |

| Location | 15,927,424 – 15,927,539 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -24.72 |
| Energy contribution | -26.16 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.92 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.999176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
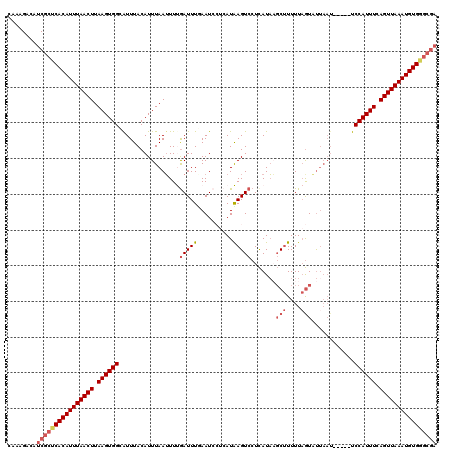
>X_DroMel_CAF1 15927424 115 + 22224390 CAAAGACAUCGCUCACAUUUAACUUAAGUGGGAUUUACAUUUAAUUUUGAUUUUAAUCCUCAUAAGUCCUCAUAAGCUUUUUAGUAUUAAU-----UCCAUUUUAGUUAAAUGUGGGCGA ........((((((((((((((((.(((((((((((((..........(((....))).....((((........))))....)))..)))-----))))))).)))))))))))))))) ( -34.60) >DroSec_CAF1 25992 112 + 1 CAAAGACAUCGCUCACAUUUAACUUAAGUGGCGUUUACAUUUAAUUUUGAUUUGAAUCCUCAUAAGUCCUCAUAAGCUUUUUAGUAUA--A-----UCCAUUU-AGUUAAAUGUGAGCGA ........((((((((((((((((.((((((....(((((((((.......))))))......((((........))))....)))..--.-----.))))))-)))))))))))))))) ( -32.80) >DroSim_CAF1 11164 115 + 1 CAAAGACAUCGCUCACAUUUAACUUAAGUGGCGUUUACAUUUAAUUUUGAUUUGAAUCCUCAUAAGUCCUCAUAAGCUUUUUAGUAUUAAU-----UCCAUUUCAGUUAAAUGUGGGCGA ........((((((((((((((((.((((((.((((((((((((.......))))))......((((........))))....)))..)))-----.)))))).)))))))))))))))) ( -32.40) >DroEre_CAF1 19394 116 + 1 CAAAGACAUCGCUCACAUUUAACUUAAGUGGCAUUUACAUUUAAUUUCGAUUUUAAU----AUGAGUCCUUAUACUCAUUUUCAUUUUAAUAUUAAUCCAUUUCAGUUAAAUGUGGGCGA ........((((((((((((((((.((((((.(((.....((((..(.((.......----((((((......))))))..)))..))))....))))))))).)))))))))))))))) ( -34.50) >DroYak_CAF1 10371 106 + 1 CAAAGACAUCGCUCACAUUUAACUUAAGUGGCAUUUACAUUUAAGUUCAAUUUCAAU----AUGAGUCCUCAUCUGCUUUUUAGUUCUAAU-----UCCAUUUUAGUUAAAUGUG----- .............(((((((((((.((((((.(((.((....((((...........----((((....))))..))))....))...)))-----.)))))).)))))))))))----- ( -20.72) >consensus CAAAGACAUCGCUCACAUUUAACUUAAGUGGCAUUUACAUUUAAUUUUGAUUUGAAUCCUCAUAAGUCCUCAUAAGCUUUUUAGUAUUAAU_____UCCAUUUCAGUUAAAUGUGGGCGA ........((((((((((((((((.((((((.................(((((..........))))).......(((....)))............)))))).)))))))))))))))) (-24.72 = -26.16 + 1.44)

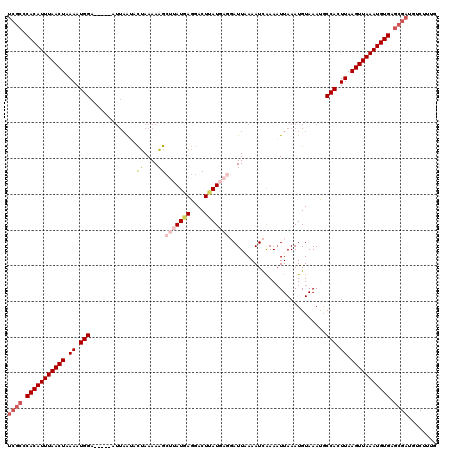
| Location | 15,927,424 – 15,927,539 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -19.42 |
| Energy contribution | -21.02 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15927424 115 - 22224390 UCGCCCACAUUUAACUAAAAUGGA-----AUUAAUACUAAAAAGCUUAUGAGGACUUAUGAGGAUUAAAAUCAAAAUUAAAUGUAAAUCCCACUUAAGUUAAAUGUGAGCGAUGUCUUUG ((((.(((((((((((.((.(((.-----(((.(((........(((((((....)))))))(((....))).........))).))).))).)).))))))))))).))))........ ( -28.80) >DroSec_CAF1 25992 112 - 1 UCGCUCACAUUUAACU-AAAUGGA-----U--UAUACUAAAAAGCUUAUGAGGACUUAUGAGGAUUCAAAUCAAAAUUAAAUGUAAACGCCACUUAAGUUAAAUGUGAGCGAUGUCUUUG ((((((((((((((((-((.(((.-----.--..(((.......(((((((....)))))))(((....)))..........)))....))).)).))))))))))))))))........ ( -33.70) >DroSim_CAF1 11164 115 - 1 UCGCCCACAUUUAACUGAAAUGGA-----AUUAAUACUAAAAAGCUUAUGAGGACUUAUGAGGAUUCAAAUCAAAAUUAAAUGUAAACGCCACUUAAGUUAAAUGUGAGCGAUGUCUUUG ((((.(((((((((((.((.(((.-----.....(((.......(((((((....)))))))(((....)))..........)))....))).)).))))))))))).))))........ ( -28.50) >DroEre_CAF1 19394 116 - 1 UCGCCCACAUUUAACUGAAAUGGAUUAAUAUUAAAAUGAAAAUGAGUAUAAGGACUCAU----AUUAAAAUCGAAAUUAAAUGUAAAUGCCACUUAAGUUAAAUGUGAGCGAUGUCUUUG ((((.(((((((((((.((.((((((.(((((.........((((((......))))))----..(((........)))))))).))).))).)).))))))))))).))))........ ( -28.90) >DroYak_CAF1 10371 106 - 1 -----CACAUUUAACUAAAAUGGA-----AUUAGAACUAAAAAGCAGAUGAGGACUCAU----AUUGAAAUUGAACUUAAAUGUAAAUGCCACUUAAGUUAAAUGUGAGCGAUGUCUUUG -----(((((((((((.((.(((.-----.(((....)))...(((..((((...(((.----........))).))))..))).....))).)).)))))))))))............. ( -19.80) >consensus UCGCCCACAUUUAACUAAAAUGGA_____AUUAAUACUAAAAAGCUUAUGAGGACUUAUGAGGAUUAAAAUCAAAAUUAAAUGUAAAUGCCACUUAAGUUAAAUGUGAGCGAUGUCUUUG ((((.(((((((((((.((.(((.............((....))(((((((....)))))))...........................))).)).))))))))))).))))........ (-19.42 = -21.02 + 1.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:48 2006