
| Sequence ID | X_DroMel_CAF1 |
|---|---|
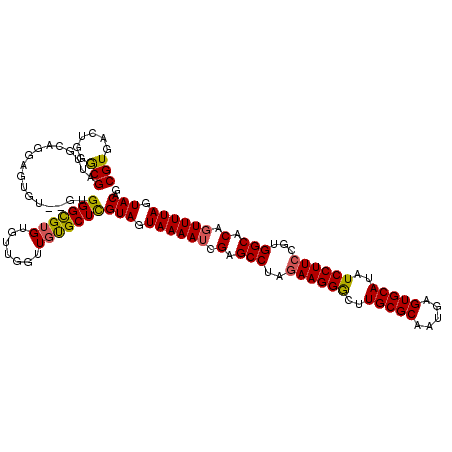
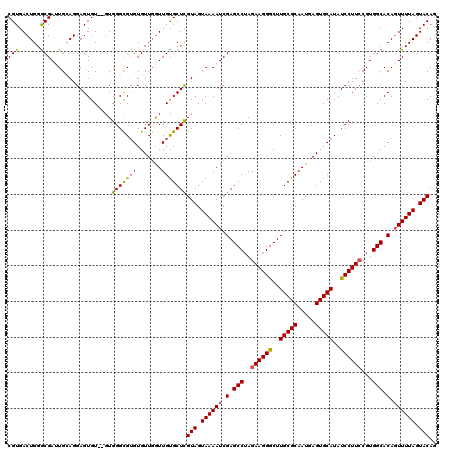
| Location | 15,926,506 – 15,926,664 |
| Length | 158 |
| Max. P | 0.819005 |

| Location | 15,926,506 – 15,926,624 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -41.24 |
| Consensus MFE | -33.16 |
| Energy contribution | -32.96 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15926506 118 + 22224390 CGUGACUGGACGAUUGGAGGAGUGU--GUGGGCGUGUGUUGGUUGUGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACAGUUUUAGUACAG .......(.(((((..(....(..(--(....))..).)..))))).)..(((.((((((.(.(((..((((((..(((((.....)))))..))))))...))).).)))))).))).. ( -38.00) >DroSec_CAF1 25127 118 + 1 CGUGACUGGGCGAUUGCAGGAGUGU--GUGGGUGUGUGUUGGUUGUGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACAGUUUUAGUACAG .(((...((((...(((.((((...--..(((((((..((.(((((((.....(......)((((((....))))))))))))).))..)))))))))))...)))..))))...))).. ( -41.10) >DroSim_CAF1 10298 120 + 1 CGUGACUGGGCGAUUGGAGGAGUGUGUGUGGGCGUGUGUUGGUUGUGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACAGUUUUAGUACAG ...(((((.((.((.((((((((.(....).))(((..((.(((((((.....(......)((((((....))))))))))))).))..))).)))))).)).)).)))))......... ( -40.70) >DroEre_CAF1 18489 118 + 1 CGUGACUGGGCGAGUGCAGGAGUGU--GUGGGCGUGUGUUUAUUGCGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUGCGUGGCACAGUUUUAGUACAG .(((...((((..((((....(((.--..((..(((..((((((((((.....(......)((((((....))))))))))))))))..)))..)).)))...)))).))))...))).. ( -42.80) >DroYak_CAF1 9480 118 + 1 CGUGACUGGGCGAGUGCAGGAGUGU--GUGGGCGAGUGUUGGUUGUGCUUGUAGUAAAAUCGAGCCAAGAAGGACUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACACUUUUAGUACAC (((......))).((((((((((((--....(((((..(.....)..)))))...........((((.((((((..(((((.....)))))..))))))..)))))))))))).)))).. ( -43.60) >consensus CGUGACUGGGCGAUUGCAGGAGUGU__GUGGGCGUGUGUUGGUUGUGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACAGUUUUAGUACAG (((......))).................(((((((.......)))))))(((.((((((.(.(((..((((((..(((((.....)))))..))))))...))).).)))))).))).. (-33.16 = -32.96 + -0.20)

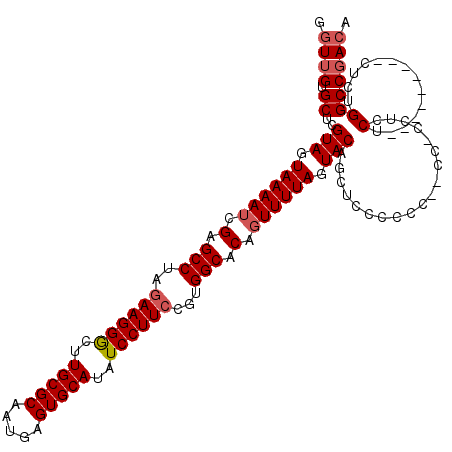
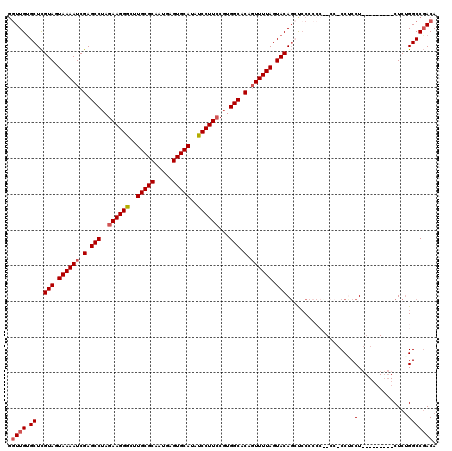
| Location | 15,926,544 – 15,926,664 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.51 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -28.78 |
| Energy contribution | -29.42 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.819005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15926544 120 + 22224390 GGUUGUGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACAGUUUUAGUACAGCCCCCCCCCUCCUCCUCCUCUUCCACCACUCUGGCCGACA .((((.(((.(((.((((((.(.(((..((((((..(((((.....)))))..))))))...))).).)))))).)))))).....................(((......))).)))). ( -35.20) >DroSec_CAF1 25165 109 + 1 GGUUGUGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACAGUUUUAGUACAGCUCCCCCCU-CC-CCUCCU---------CUCUGGCCGACA .((((.(((.(((.((((((.(.(((..((((((..(((((.....)))))..))))))...))).).)))))).))))))........-..-((....---------....)).)))). ( -35.30) >DroSim_CAF1 10338 109 + 1 GGUUGUGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACAGUUUUAGUACAGCUCCCCCCU-CC-CCUCCU---------CUCGGGCCGACA .((((.(((.(((.((((((.(.(((..((((((..(((((.....)))))..))))))...))).).)))))).))))))........-.(-((....---------...))).)))). ( -37.70) >DroEre_CAF1 18527 102 + 1 UAUUGCGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUGCGUGGCACAGUUUUAGUACAGAUCC---------CCGCCU---------CUCUGGCCGACA ((((((....))))))...(((.(((.(((..((((..(((((.((.......)).)))))..)....((......))......---------..))).---------.))))))))).. ( -30.80) >DroYak_CAF1 9518 102 + 1 GGUUGUGCUUGUAGUAAAAUCGAGCCAAGAAGGACUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACACUUUUAGUACACAUGC---------CCGCCU---------CUCUGGCCCACA ((..(((..((((.(((((..(.((((.((((((..(((((.....)))))..))))))..)))).)..))))).)))))))..---------))(((.---------....)))..... ( -37.90) >consensus GGUUGUGCUCGUAGUAAAAUCGAGCCUAGAAGGGCUUGCGCAAUGAGUGCAUAUCCUUCCGUGGCACAGUUUUAGUACAGCUCCCCCC__CC_CCUCCU_________CUCUGGCCGACA .((((.((..(((.((((((.(.(((..((((((..(((((.....)))))..))))))...))).).)))))).)))...................(..............))))))). (-28.78 = -29.42 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:46 2006