| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,924,565 – 15,924,667 |
| Length | 102 |
| Max. P | 0.995213 |

| Location | 15,924,565 – 15,924,667 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.44 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -6.53 |
| Energy contribution | -7.18 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.22 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
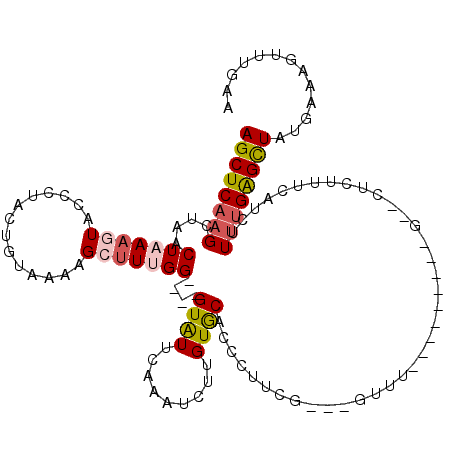
>X_DroMel_CAF1 15924565 102 + 22224390 AGCUCAAGCCAACUAAAGUACCCUACUGUAAAAGCUUUGG----GUAUUCAAAUCUUGUGCACCCUUCG---GUUU---------G--CUCUUUCAUCUUUUGAGCUACGAAAGUUUGAA ((((((((........(((((((....((....))...))----)))))........(.(((..(....---)..)---------)--).)........))))))))((....))..... ( -24.70) >DroSec_CAF1 23306 101 + 1 AGCUCAAGCUAACUAAAGUACCCUACUGUAAAAGCUUUGG----GUAUUCAAAUCUUGUACACCCUUCG----UUA---------G--CUCUUUCAUCUUGUGAGCUAUGAAAGUUUGAA ...(((((((......(((((((....((....))...))----)))))................((((----(.(---------(--(((...........))))))))))))))))). ( -24.40) >DroSim_CAF1 7993 85 + 1 AGCUCA-GCUA-CU-AAGUAC-CUACUGUAA-AGCUUUGG----GUAUC--AAUC-UGUGCA--CUUCG-----UA---------G--CUCU-UCAUCUU-UGAGCUAUGAAG---UCGA .((.((-(...-..-..((((-(((..(...-..)..)))----)))).--...)-)).))(--(((((-----((---------(--(((.-.......-.)))))))))))---)... ( -29.20) >DroEre_CAF1 16711 118 + 1 AGCUCAAGCCAACUAAAGUACCCUACAGUAAGUGGAUUGGGUGGGUGUUCAAAUUUUGUGCACCCUUCGUUAGUUUGAGUGCCGAG--CCCUUUCAUAAUUUGGGUUAUGAAAGUUCGAA .((((((((.(((......((((..((.....))....))))((((((.((.....)).))))))...))).))))))))..((((--(..(((((((((....)))))))))))))).. ( -39.50) >DroYak_CAF1 7904 114 + 1 AGCUCAAGCUAACUUAAGUUGUCUAUAGUAAGAGGUUUGG----GUGUUGAAAUGUUGUGCCCUCUUCGU--GCUUGAGUGCCGAGAACCCCUUCAUAUUUGGAGUUAUGAAAGUUCGAA .((((((((((((....))))..........((((...((----(..(.........)..))).))))..--)))))))).....((((...((((((........)))))).))))... ( -30.50) >consensus AGCUCAAGCUAACUAAAGUACCCUACUGUAAAAGCUUUGG____GUAUUCAAAUCUUGUGCACCCUUCG___GUUU_________G__CUCUUUCAUCUUUUGAGCUAUGAAAGUUUGAA ((((((((....(((((((..............)))))))....((((.........))))......................................))))))))............. ( -6.53 = -7.18 + 0.65)


| Location | 15,924,565 – 15,924,667 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.44 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -4.78 |
| Energy contribution | -4.86 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.18 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
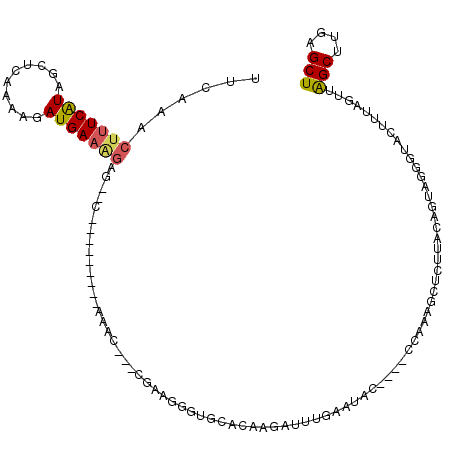
>X_DroMel_CAF1 15924565 102 - 22224390 UUCAAACUUUCGUAGCUCAAAAGAUGAAAGAG--C---------AAAC---CGAAGGGUGCACAAGAUUUGAAUAC----CCAAAGCUUUUACAGUAGGGUACUUUAGUUGGCUUGAGCU ......((((((..((((...........)))--)---------....---))))))..((.((((..((((((((----((...(((.....))).))))).)))))....)))).)). ( -24.30) >DroSec_CAF1 23306 101 - 1 UUCAAACUUUCAUAGCUCACAAGAUGAAAGAG--C---------UAA----CGAAGGGUGUACAAGAUUUGAAUAC----CCAAAGCUUUUACAGUAGGGUACUUUAGUUAGCUUGAGCU .............(((((.(.....)...(((--(---------(((----(...((((((.((.....)).))))----))...(((((......)))))......))))))))))))) ( -26.10) >DroSim_CAF1 7993 85 - 1 UCGA---CUUCAUAGCUCA-AAGAUGA-AGAG--C---------UA-----CGAAG--UGCACA-GAUU--GAUAC----CCAAAGCU-UUACAGUAG-GUACUU-AG-UAGC-UGAGCU ...(---((((.((((((.-.......-.)))--)---------))-----.))))--)((.((-((((--(((((----(....(((-....))).)-))).))-))-)..)-)).)). ( -22.10) >DroEre_CAF1 16711 118 - 1 UUCGAACUUUCAUAACCCAAAUUAUGAAAGGG--CUCGGCACUCAAACUAACGAAGGGUGCACAAAAUUUGAACACCCACCCAAUCCACUUACUGUAGGGUACUUUAGUUGGCUUGAGCU ......(((((((((......)))))))))((--((((((.....((((((.(..(((((..((.....))..)))))((((...............)))).).)))))).)).)))))) ( -34.26) >DroYak_CAF1 7904 114 - 1 UUCGAACUUUCAUAACUCCAAAUAUGAAGGGGUUCUCGGCACUCAAGC--ACGAAGAGGGCACAACAUUUCAACAC----CCAAACCUCUUACUAUAGACAACUUAAGUUAGCUUGAGCU .((((.((((((((........)))))))).....))))..(((((((--...((((((.................----.....)))))).........(((....))).))))))).. ( -25.45) >consensus UUCAAACUUUCAUAGCUCAAAAGAUGAAAGAG__C_________AAAC___CGAAGGGUGCACAAGAUUUGAAUAC____CCAAAGCUCUUACAGUAGGGUACUUUAGUUAGCUUGAGCU ......(((((((..........)))))))................................................................................(((....))) ( -4.78 = -4.86 + 0.08)

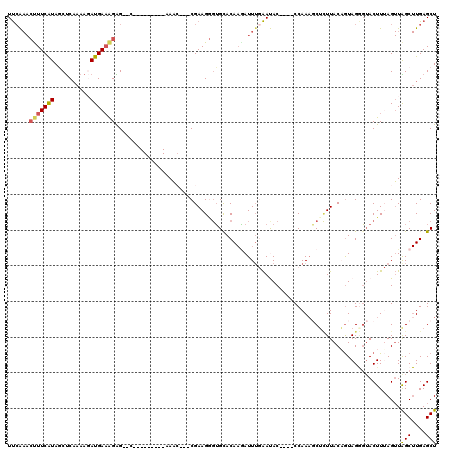
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:44 2006