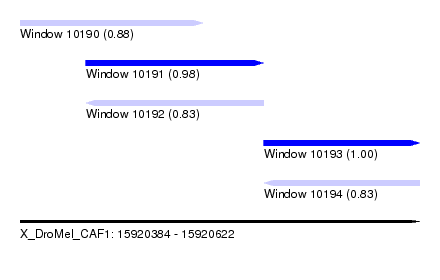
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,920,384 – 15,920,622 |
| Length | 238 |
| Max. P | 0.995003 |

| Location | 15,920,384 – 15,920,493 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.13 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -17.28 |
| Energy contribution | -20.44 |
| Covariance contribution | 3.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15920384 109 + 22224390 UUAUUUUGAAUUUU-AGUAGAAGAGAAAGCCAUUUCUCUUUUCUUUCUCUGAUUUUCACUCACUUUUCUCCAGAGUGAGUGGGUCUCAGUGCGCAGUAACAUAACAU----------ACA (((((.((......-((.((((((((((....))))))))))))..(.((((..((((((((((((.....))))))))))))..)))).))).)))))........----------... ( -29.80) >DroSec_CAF1 18915 109 + 1 UUAUUUUGAAUUUU-AGUAGAAGAGAAAGCAAUUUCUCUUGUCUUUCUCUGAUUCUCACUCACUUUUCUCCAGAGUGAGUGGGUCCUAGUGCGCAGUAACAUAACAU----------ACA (((((.((....((-((....(((((((((((......))).))))))))....((((((((((((.....))))))))))))..))))..)).)))))........----------... ( -29.10) >DroSim_CAF1 3732 109 + 1 CUAUUUUCAAUAUU-AAUAGAAGAGAAAGCAAUUUCUCUUUUCUUUCUCUGAUUUUCACUCACUUUUCUCCAGAGUGAGUGGGUCCCAGUGCGCAGUAACAUAACAU----------ACA ..............-...((((((((((....))))))))))....(.(((...((((((((((((.....))))))))))))...))).)....(((........)----------)). ( -27.10) >DroEre_CAF1 12621 107 + 1 UUAUUCAGCAUUUU-AGUA-----------GAAUUAUCUUUU-UUUCUCUGAUUUUCACUCACUUUUCUCCAGAGUGCGUGGGUCCCAGUGCGCAGUAACAUAACAUACAUGUACAUACA .......(((((((-((.(-----------(((..(....).-.))))))))..(((((.((((((.....)))))).)))))....)))))((((((........))).)))....... ( -19.10) >DroYak_CAF1 3707 110 + 1 UUAUUUUGAAUUUUUAUUAGAAGAGCAAGCGAAUUCUCUUUUCUUUCUCUGAUUUCCACUCACGUUUCUCCAGAGUGAGUGGCUCCCAGUGCGCAGUAACAUAACAU----------ACG (((((.((..........(((((((.((.....)))))))))....(.(((....((((((((...........))))))))....))).))).)))))........----------... ( -19.50) >consensus UUAUUUUGAAUUUU_AGUAGAAGAGAAAGCAAUUUCUCUUUUCUUUCUCUGAUUUUCACUCACUUUUCUCCAGAGUGAGUGGGUCCCAGUGCGCAGUAACAUAACAU__________ACA ..................((((((((((....))))))))))....(.(((...((((((((((((.....))))))))))))...))).)............................. (-17.28 = -20.44 + 3.16)


| Location | 15,920,423 – 15,920,529 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.49 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15920423 106 + 22224390 UUCUUUCUCUGAUUUUCACUCACUUUUCUCCAGAGUGAGUGGGUCUCAGUGCGCAGUAACAUAACAU----------ACAU----ACGUCACAGAGACCUCAGCGCCCGCCCACACACAU ........((((....((((((((((.....))))))))))((((((.(((((..(((........)----------))..----.)).))).))))))))))................. ( -30.40) >DroSec_CAF1 18954 108 + 1 GUCUUUCUCUGAUUCUCACUCACUUUUCUCCAGAGUGAGUGGGUCCUAGUGCGCAGUAACAUAACAU----------ACAUAUGUACGUCACAGAGACCUCAGC--CCGCCCACACACAU ........((((....((((((((((.....))))))))))((((((.(((.((.(((.((((....----------...)))))))))))))).)))))))).--.............. ( -28.60) >DroSim_CAF1 3771 104 + 1 UUCUUUCUCUGAUUUUCACUCACUUUUCUCCAGAGUGAGUGGGUCCCAGUGCGCAGUAACAUAACAU----------ACAU----ACGUCACAGAGACCUCAGC--CCGCCCACACACAC ........((((....((((((((((.....))))))))))((((.(.(((((..(((........)----------))..----.)).))).).)))))))).--.............. ( -25.60) >DroEre_CAF1 12649 113 + 1 UU-UUUCUCUGAUUUUCACUCACUUUUCUCCAGAGUGCGUGGGUCCCAGUGCGCAGUAACAUAACAUACAUGUACAUACAU----ACGUCACAGAGACCUCAGC--CCGCCCACACAAAU ..-.....((((....(((.((((((.....)))))).)))((((.(.(((((..(((........)))((((....))))----.)).))).).)))))))).--.............. ( -22.40) >DroYak_CAF1 3747 104 + 1 UUCUUUCUCUGAUUUCCACUCACGUUUCUCCAGAGUGAGUGGCUCCCAGUGCGCAGUAACAUAACAU----------ACGU----ACGUCACAGAGACCUCAGU--CCGCCCACACACAU .....((((((....((((((((...........))))))))......(((((..((......))..----------.)))----))....)))))).......--.............. ( -25.80) >consensus UUCUUUCUCUGAUUUUCACUCACUUUUCUCCAGAGUGAGUGGGUCCCAGUGCGCAGUAACAUAACAU__________ACAU____ACGUCACAGAGACCUCAGC__CCGCCCACACACAU .....((((((...((((((((((((.....)))))))))))).....((........))...............................))))))....................... (-20.70 = -21.02 + 0.32)


| Location | 15,920,423 – 15,920,529 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.49 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15920423 106 - 22224390 AUGUGUGUGGGCGGGCGCUGAGGUCUCUGUGACGU----AUGU----------AUGUUAUGUUACUGCGCACUGAGACCCACUCACUCUGGAGAAAAGUGAGUGAAAAUCAGAGAAAGAA ..((.((....)).)).((((((((((.(((.(((----(.((----------(........)))))))))).))))))((((((((.(.....).))))))))....))))........ ( -37.30) >DroSec_CAF1 18954 108 - 1 AUGUGUGUGGGCGG--GCUGAGGUCUCUGUGACGUACAUAUGU----------AUGUUAUGUUACUGCGCACUAGGACCCACUCACUCUGGAGAAAAGUGAGUGAGAAUCAGAGAAAGAC ..(((((..(..((--((....))))..(((((((((....))----------)))))))....)..))))).......((((((((.(.....).))))))))................ ( -32.70) >DroSim_CAF1 3771 104 - 1 GUGUGUGUGGGCGG--GCUGAGGUCUCUGUGACGU----AUGU----------AUGUUAUGUUACUGCGCACUGGGACCCACUCACUCUGGAGAAAAGUGAGUGAAAAUCAGAGAAAGAA ..............--.((((((((((.(((.(((----(.((----------(........)))))))))).))))))((((((((.(.....).))))))))....))))........ ( -33.80) >DroEre_CAF1 12649 113 - 1 AUUUGUGUGGGCGG--GCUGAGGUCUCUGUGACGU----AUGUAUGUACAUGUAUGUUAUGUUACUGCGCACUGGGACCCACGCACUCUGGAGAAAAGUGAGUGAAAAUCAGAGAAA-AA ...((((((((...--.))..((((((.(((.(((----(.(((...(((((.....))))))))))))))).))))))))))))((((((.................))))))...-.. ( -32.93) >DroYak_CAF1 3747 104 - 1 AUGUGUGUGGGCGG--ACUGAGGUCUCUGUGACGU----ACGU----------AUGUUAUGUUACUGCGCACUGGGAGCCACUCACUCUGGAGAAACGUGAGUGGAAAUCAGAGAAAGAA ...((((..(..((--((....))))..(((((((----....----------)))))))....)..))))((((...((((((((...........))))))))...))))........ ( -32.60) >consensus AUGUGUGUGGGCGG__GCUGAGGUCUCUGUGACGU____AUGU__________AUGUUAUGUUACUGCGCACUGGGACCCACUCACUCUGGAGAAAAGUGAGUGAAAAUCAGAGAAAGAA .................((((((((((.(((.(((....((((...............))))....)))))).))))))((((((((.(.....).))))))))....))))........ (-26.16 = -26.64 + 0.48)


| Location | 15,920,529 – 15,920,622 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -29.27 |
| Energy contribution | -29.53 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15920529 93 + 22224390 A---UUUUUUAGCACCAGUGCAGUUGUGUUGAUGCAGCGGGGUCAUGCGUGCUAGUAUGCGUAUCUAUUGCAUGUACAUAACCUGCUUUUUGCAUU .---.......(((.....((((((((((..((((((..((((.(((((((....))))))))))).))))))..)))))).))))....)))... ( -30.40) >DroSec_CAF1 19062 93 + 1 A---UUUUUUAGCACCAGUGCAGUUGUGUUGAUGCAGCGGGGUCAUGCGUGCUUGUAUGCGUAUCUAUUGCAUGUACAUAACCUGCUUUUUGCAUU .---.......(((.....((((((((((..((((((..((((.(((((((....))))))))))).))))))..)))))).))))....)))... ( -30.40) >DroSim_CAF1 3875 95 + 1 AUU-UUUUUUAGCACCAGUGCAGUUGUGUUGAUGCAGCGGGGUCAUGCGUGCUUGUAUGCGUAUCUAUUGCAUGUACAUAACCUGCUUUUUGCAUU ...-.......(((.....((((((((((..((((((..((((.(((((((....))))))))))).))))))..)))))).))))....)))... ( -30.40) >DroEre_CAF1 12762 96 + 1 AUUUUUUUUUAGCACUAGUGCAGCUGUGUUGAUGCAGCGGGGUCAUGCGUGUUUGUAUGCGUAUCUAAUGCAUGUACAUAACCUGCUUUUUGCAUU ...........(((..((.((((.(((((..(((((...((((.(((((((....)))))))))))..)))))..)))))..))))))..)))... ( -28.20) >consensus A___UUUUUUAGCACCAGUGCAGUUGUGUUGAUGCAGCGGGGUCAUGCGUGCUUGUAUGCGUAUCUAUUGCAUGUACAUAACCUGCUUUUUGCAUU ...........(((.....((((((((((..(((((...((((.(((((((....)))))))))))..)))))..)))))).))))....)))... (-29.27 = -29.53 + 0.25)



| Location | 15,920,529 – 15,920,622 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -17.17 |
| Energy contribution | -17.43 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15920529 93 - 22224390 AAUGCAAAAAGCAGGUUAUGUACAUGCAAUAGAUACGCAUACUAGCACGCAUGACCCCGCUGCAUCAACACAACUGCACUGGUGCUAAAAAA---U .........((((((((((((...(((..(((.........)))))).))))))))(((.((((..........)))).)))))))......---. ( -20.00) >DroSec_CAF1 19062 93 - 1 AAUGCAAAAAGCAGGUUAUGUACAUGCAAUAGAUACGCAUACAAGCACGCAUGACCCCGCUGCAUCAACACAACUGCACUGGUGCUAAAAAA---U .........((((((((((((...(((.................))).))))))))(((.((((..........)))).)))))))......---. ( -19.63) >DroSim_CAF1 3875 95 - 1 AAUGCAAAAAGCAGGUUAUGUACAUGCAAUAGAUACGCAUACAAGCACGCAUGACCCCGCUGCAUCAACACAACUGCACUGGUGCUAAAAAA-AAU .........((((((((((((...(((.................))).))))))))(((.((((..........)))).)))))))......-... ( -19.63) >DroEre_CAF1 12762 96 - 1 AAUGCAAAAAGCAGGUUAUGUACAUGCAUUAGAUACGCAUACAAACACGCAUGACCCCGCUGCAUCAACACAGCUGCACUAGUGCUAAAAAAAAAU ...(((....(((((((((((..((((.........))))........))))))))..((((........))))))).....)))........... ( -19.40) >consensus AAUGCAAAAAGCAGGUUAUGUACAUGCAAUAGAUACGCAUACAAGCACGCAUGACCCCGCUGCAUCAACACAACUGCACUGGUGCUAAAAAA___U .........((((((((((((..((((.........))))........))))))))(((.((((..........)))).))))))).......... (-17.17 = -17.43 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:37 2006