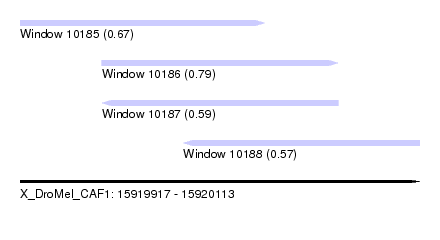
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,919,917 – 15,920,113 |
| Length | 196 |
| Max. P | 0.789537 |

| Location | 15,919,917 – 15,920,037 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.30 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -32.26 |
| Energy contribution | -31.82 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15919917 120 + 22224390 CACUGCAUUCGGGAAUGAAACUGGAGCACAGCUGAGCGCUAGGCCGAAGUGGGACGAAACACCCACAUAAAGGUAAUGGGCGCUAUCGAGAGAGCGGGAGAGUAAGACCACGUUCUAAUC ...(((.(((((........))))))))......((((((..(((...(((((........))))).....)))....))))))......((((((((.........)).)))))).... ( -36.90) >DroSec_CAF1 18467 118 + 1 CACUGCAUUCGGGAACGAAACUGGAGCACAGCUGAGUGCUAGGCCAAAGUGGGACGA-ACACCCACAUAACGGUAACGGGCGCUAUCGAGAGAGUGGGAGAGCAAGACCAUGUUCUAAA- ((((...((((....)))).((.((((((......))))((((((...(((((....-...)))))....((....))))).))))).))..))))..((((((......))))))...- ( -37.70) >DroSim_CAF1 3283 118 + 1 CACUGCAUUCGGGAAUGAAACUGGAGCACAGCUGAGUGCUAGGCCGAAGUGGGACGA-ACACCCACAUAACGGUAACGGGCGCUAUCGAGAGAGUGGGAGAGUAAGACCAUGUUCUAAA- ...(((.(((((........))))))))......((((((..((((..(((((....-...)))))....))))....))))))....((((.((((..........)))).))))...- ( -34.90) >DroYak_CAF1 3308 118 + 1 CAAUGCAUUCGGGAGUGAAACUGGAGCACAGCUGAGCGGUAGGCCGAAGUGGGACGA-ACACCCACAUAACGGUAACGGCCGCUAUCGAGAGAGUGGAAGAGUAAGACAAUAUUCUAAA- ...(((.(((((........))))))))......((((((..((((..(((((....-...)))))....))))....)))))).((....)).....((((((......))))))...- ( -35.40) >consensus CACUGCAUUCGGGAAUGAAACUGGAGCACAGCUGAGCGCUAGGCCGAAGUGGGACGA_ACACCCACAUAACGGUAACGGGCGCUAUCGAGAGAGUGGGAGAGUAAGACCAUGUUCUAAA_ ...(((.(((((........))))))))..((..((((((..((((..(((((........)))))....))))....)))))).((....))))...((((((......)))))).... (-32.26 = -31.82 + -0.44)

| Location | 15,919,957 – 15,920,073 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.53 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -28.92 |
| Energy contribution | -29.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15919957 116 + 22224390 AGGCCGAAGUGGGACGAAACACCCACAUAAAGGUAAUGGGCGCUAUCGAGAGAGCGGGAGAGUAAGACCACGUUCUAAUCCGCUCUCCACGC--UCCACGCUCUACGCAGCUGAUCCG ..(((...(((((........))))).....))).((.(((((....(((.((((((((((((.(((......))).....))))))).)))--))....)))...)).))).))... ( -37.60) >DroSec_CAF1 18507 107 + 1 AGGCCAAAGUGGGACGA-ACACCCACAUAACGGUAACGGGCGCUAUCGAGAGAGUGGGAGAGCAAGACCAUGUUCUAAA-CGCUCUC---------CACGCUCUACGCAGCUGAUCCG ..(((...(((((....-...)))))....((....)))))(((..((..(((((((((((((.(((......)))...-.))))))---------).)))))).)).)))....... ( -39.50) >DroSim_CAF1 3323 107 + 1 AGGCCGAAGUGGGACGA-ACACCCACAUAACGGUAACGGGCGCUAUCGAGAGAGUGGGAGAGUAAGACCAUGUUCUAAA-CGCUCUC---------CACGCUCUACGCAGCUGAUCCG ..((((..(((((....-...)))))....))))..((((((((..((..(((((((((((((.(((......)))...-.))))))---------).)))))).)).))).).)))) ( -38.80) >DroYak_CAF1 3348 116 + 1 AGGCCGAAGUGGGACGA-ACACCCACAUAACGGUAACGGCCGCUAUCGAGAGAGUGGAAGAGUAAGACAAUAUUCUAAA-CGCUCUCCACGCUCUCCACGCUUUACGCAGCUGGUCCG ..((((..(((((....-...)))))....))))..((((((((...(((((.(((((.((((.(((......)))...-.))))))))).)))))...((.....))))).)).))) ( -42.30) >consensus AGGCCGAAGUGGGACGA_ACACCCACAUAACGGUAACGGGCGCUAUCGAGAGAGUGGGAGAGUAAGACCAUGUUCUAAA_CGCUCUC_________CACGCUCUACGCAGCUGAUCCG ..((((..(((((........)))))....))))..((((((((.....(((((((..((((((......))))))....)))))))............((.....))))).).)))) (-28.92 = -29.18 + 0.25)



| Location | 15,919,957 – 15,920,073 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 88.53 |
| Mean single sequence MFE | -41.42 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15919957 116 - 22224390 CGGAUCAGCUGCGUAGAGCGUGGA--GCGUGGAGAGCGGAUUAGAACGUGGUCUUACUCUCCCGCUCUCUCGAUAGCGCCCAUUACCUUUAUGUGGGUGUUUCGUCCCACUUCGGCCU .((.((.(((......)))(((((--(((.((((((.((((((.....))))))..)))))))))))...(((.(((((((((.........))))))))))))...)))...)))). ( -44.00) >DroSec_CAF1 18507 107 - 1 CGGAUCAGCUGCGUAGAGCGUG---------GAGAGCG-UUUAGAACAUGGUCUUGCUCUCCCACUCUCUCGAUAGCGCCCGUUACCGUUAUGUGGGUGU-UCGUCCCACUUUGGCCU (((....((((((.((((.(((---------(((((((-...(((......))))))))).)))).)))))).))))..)))...(((....(((((...-....)))))..)))... ( -39.80) >DroSim_CAF1 3323 107 - 1 CGGAUCAGCUGCGUAGAGCGUG---------GAGAGCG-UUUAGAACAUGGUCUUACUCUCCCACUCUCUCGAUAGCGCCCGUUACCGUUAUGUGGGUGU-UCGUCCCACUUCGGCCU (((....((((((.((((.(((---------(.(((.(-(..(((......))).)).))))))).)))))).))))..)))...(((....(((((...-....)))))..)))... ( -39.20) >DroYak_CAF1 3348 116 - 1 CGGACCAGCUGCGUAAAGCGUGGAGAGCGUGGAGAGCG-UUUAGAAUAUUGUCUUACUCUUCCACUCUCUCGAUAGCGGCCGUUACCGUUAUGUGGGUGU-UCGUCCCACUUCGGCCU (((.((.(((((.....))((.(((((.((((((((..-...(((......)))..))).))))).))))).))))))))))...(((....(((((...-....)))))..)))... ( -42.70) >consensus CGGAUCAGCUGCGUAGAGCGUG_________GAGAGCG_UUUAGAACAUGGUCUUACUCUCCCACUCUCUCGAUAGCGCCCGUUACCGUUAUGUGGGUGU_UCGUCCCACUUCGGCCU .((....((((((.((((.((((........(((((......(((......)))..))))))))).)))))).))))..))....(((....(((((........)))))..)))... (-30.00 = -30.50 + 0.50)


| Location | 15,919,997 – 15,920,113 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15919997 116 - 22224390 UUCCACAUCCGAAGGCCAAGAAAUGCGCUUCGUGUGUCCUCGGAUCAGCUGCGUAGAGCGUGGA--GCGUGGAGAGCGGAUUAGAACGUGGUCUUACUCUCCCGCUCUCUCGAUAGCG ......((((((.(((((.(((......))).)).))).))))))..((((((.((((((.(((--(.(((((..(((........)))..)).))).))))))))))..)).)))). ( -45.00) >DroSec_CAF1 18546 108 - 1 UUCCACAUCCGAAGGCCAAGAAAUGCGCUUCGUGUGUCCUCGGAUCAGCUGCGUAGAGCGUG---------GAGAGCG-UUUAGAACAUGGUCUUGCUCUCCCACUCUCUCGAUAGCG ......((((((.(((((.(((......))).)).))).))))))..((((((.((((.(((---------(((((((-...(((......))))))))).)))).)))))).)))). ( -40.80) >DroSim_CAF1 3362 108 - 1 UUCCACAUCCGAAGGCCAAGAAAUGCGCUUCGUGUGUCCUCGGAUCAGCUGCGUAGAGCGUG---------GAGAGCG-UUUAGAACAUGGUCUUACUCUCCCACUCUCUCGAUAGCG ......((((((.(((((.(((......))).)).))).))))))..((((((.((((.(((---------(.(((.(-(..(((......))).)).))))))).)))))).)))). ( -38.50) >DroYak_CAF1 3387 117 - 1 UUCCACAUCCGAAGGCCAAGAACUGCGCUUCGUGUGUCCUCGGACCAGCUGCGUAAAGCGUGGAGAGCGUGGAGAGCG-UUUAGAAUAUUGUCUUACUCUUCCACUCUCUCGAUAGCG ...(((.(((((.(((((.(((......))).)).))).)))))...(((......))))))(((((.((((((((..-...(((......)))..))).))))).)))))....... ( -38.20) >consensus UUCCACAUCCGAAGGCCAAGAAAUGCGCUUCGUGUGUCCUCGGAUCAGCUGCGUAGAGCGUG_________GAGAGCG_UUUAGAACAUGGUCUUACUCUCCCACUCUCUCGAUAGCG .......(((((.(((((.(((......))).)).))).)))))...((((((.((((.((((........(((((......(((......)))..))))))))).)))))).)))). (-30.40 = -30.77 + 0.38)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:28 2006