| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,917,505 – 15,917,612 |
| Length | 107 |
| Max. P | 0.500000 |

| Location | 15,917,505 – 15,917,612 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -14.81 |
| Energy contribution | -16.03 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15917505 107 + 22224390 GCUUACAGGACGAUUGAGGAACGGAAAAAGUUGUGCAUAUUAAUAGCAGGACUAAAUUGUUUCUCUAGAAUUCCACCGGUAAGGGGAAAACUAUGUAAA-AU----GU-AUAU ...((((..(((...(((((((((....((((.(((.........))).))))...)))))))))(((..((((.((.....))))))..))))))...-.)----))-)... ( -26.80) >DroVir_CAF1 10183 109 + 1 GCAUUCCGCACUAUAGAGGAGCGUAAGAAGUUGUGCCUGAUUAUAUCUGGCUUGUCGUGCUUCAGUAAUAUACCGCCGGUAGGUAU-UACCUAUU-UGC-CCGAUAGU-AUAG ((.....))(((((...((.......(((((((.(((.(((...))).)))....)).)))))...........((..(((((...-..))))).-.))-)).)))))-.... ( -25.30) >DroSec_CAF1 16054 106 + 1 GCUUACAGGACGAUUGAGGAACGAAAAAAGUUGUGCAUAUUAAUAGCAGGACUAAAUUGUUUCUCUAGAAUUCCACCGGUAAGGGGAGAACUACG-AAA-AU----GU-AUAU ...((((...((...(((((((((....((((.(((.........))).))))...)))))))))(((..((((.((.....))))))..)))))-...-.)----))-)... ( -26.90) >DroSim_CAF1 884 106 + 1 GCUUACAGGACGAUCGAGGAACGAAAAAAGUUGUGCAUAUUAAUAGCAGGACUCAAUUGUUUCUCUAGAAUUCCACCGGUAAGGGGAGAACUACG-AAA-AU----GU-AUAU ...((((...((...(((((((((....((((.(((.........))).))))...)))))))))(((..((((.((.....))))))..)))))-...-.)----))-)... ( -27.20) >DroEre_CAF1 9811 106 + 1 GCUUUUAGGACAAUUGAGGAACGCAAAAAGUUGUGCAUAUUAAUAGCAGGACUAAAUUGUUUUUCUAGAAUUCCACCGGUAAGGGGAAAAUUAAA-GAA-AU----UA-AUAU ..((((((........(((((.((((..((((.(((.........))).))))...)))).)))))....((((.((.....))))))..)))))-)..-..----..-.... ( -20.40) >DroYak_CAF1 921 108 + 1 GCUUUCAGGACAAUUGAGGAACGCAAAAAGUUGUGCAUUUUAAUAGCAGGACUAAAUUGUUUCUCUAGAAUACCACCGGUAAGGGGGAAAUUACA-AAAUAU----UAUAUAU ..((((.........((((((((.....((((.(((.........))).))))....)))))))).......((.((.....)))))))).....-......----....... ( -21.30) >consensus GCUUACAGGACGAUUGAGGAACGAAAAAAGUUGUGCAUAUUAAUAGCAGGACUAAAUUGUUUCUCUAGAAUUCCACCGGUAAGGGGAAAACUACG_AAA_AU____GU_AUAU ...............(((((((((....((((.(((.........))).))))...))))))))).....((((.((.....))))))......................... (-14.81 = -16.03 + 1.22)

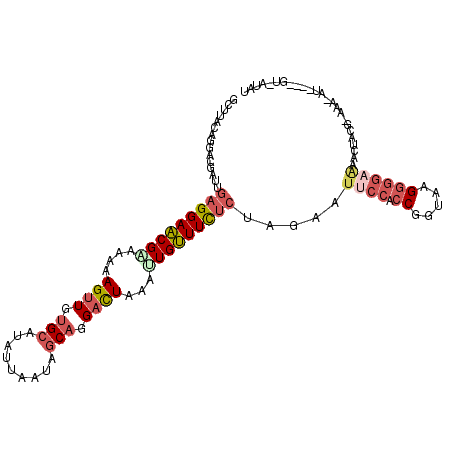

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:23 2006