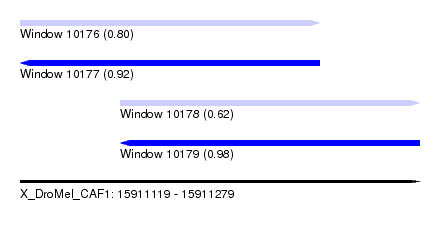
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,911,119 – 15,911,279 |
| Length | 160 |
| Max. P | 0.984204 |

| Location | 15,911,119 – 15,911,239 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -27.93 |
| Energy contribution | -29.27 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15911119 120 + 22224390 CACUUAUGCGCUACCAUGAUCAGAAUACAAAAGCUAUAGACCCAAUGCAAUCUAUAAUUCGCACCUCUUACUUUGAAGUGCGAGUGAUAUAGCAUUUAGUGGGGUAUUUUUGUUUGGCUU .........((((....(((.(((((((.....(....).((((....((((((((((((((((.((.......)).))))))))..))))).)))...))))))))))).))))))).. ( -28.30) >DroSim_CAF1 6686 118 + 1 --CUUAUGCGCUACUAUGAUCAGAAUACAAAAGCUAUAGACCCAAUGCAAUCUAUAAUUCGCACCUCUUAGUUUGAAGUGCGAGUGAUAUAGCAUUUAGUGGGGUAUUUUGGUUUGGCUU --.......((((....(((((((((((.....(....).((((....((((((((((((((((.((.......)).))))))))..))))).)))...))))))))))))))))))).. ( -33.60) >DroYak_CAF1 6877 118 + 1 --CUUAUGCGCUACUAUGAUCAGAAUACAAAAGCUAUAGACCCAAUGCAAUCUAUAAUUCGCACCUAUUACUUUGAGGUGCGAUUAAUAUAGCAUUUAAUGGGGUUUUUUGGUUUGGCUU --.......((((....((((((((.((.....(....).((((.((.((((((((..((((((((.........))))))))....))))).))))).)))))).)))))))))))).. ( -29.80) >consensus __CUUAUGCGCUACUAUGAUCAGAAUACAAAAGCUAUAGACCCAAUGCAAUCUAUAAUUCGCACCUCUUACUUUGAAGUGCGAGUGAUAUAGCAUUUAGUGGGGUAUUUUGGUUUGGCUU .........((((....(((((((((((.....(....).((((.((.((((((((((((((((.((.......)).))))))))..))))).))))).))))))))))))))))))).. (-27.93 = -29.27 + 1.33)

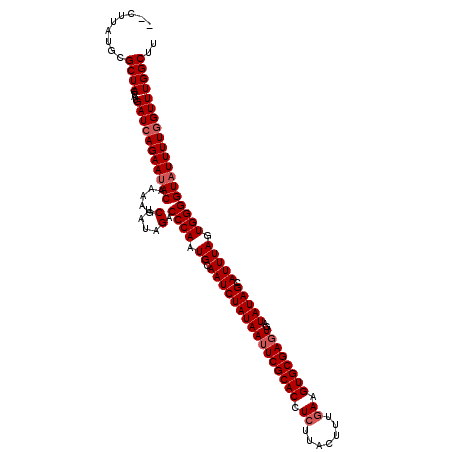

| Location | 15,911,119 – 15,911,239 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -26.59 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15911119 120 - 22224390 AAGCCAAACAAAAAUACCCCACUAAAUGCUAUAUCACUCGCACUUCAAAGUAAGAGGUGCGAAUUAUAGAUUGCAUUGGGUCUAUAGCUUUUGUAUUCUGAUCAUGGUAGCGCAUAAGUG ...................((((..((((..(((((.(((((((((.......)))))))))....((((.((((..((........))..)))).))))....)))))..)))).)))) ( -30.80) >DroSim_CAF1 6686 118 - 1 AAGCCAAACCAAAAUACCCCACUAAAUGCUAUAUCACUCGCACUUCAAACUAAGAGGUGCGAAUUAUAGAUUGCAUUGGGUCUAUAGCUUUUGUAUUCUGAUCAUAGUAGCGCAUAAG-- ...........................(((((.....(((((((((.......)))))))))....((((.((((..((........))..)))).))))......))))).......-- ( -27.00) >DroYak_CAF1 6877 118 - 1 AAGCCAAACCAAAAAACCCCAUUAAAUGCUAUAUUAAUCGCACCUCAAAGUAAUAGGUGCGAAUUAUAGAUUGCAUUGGGUCUAUAGCUUUUGUAUUCUGAUCAUAGUAGCGCAUAAG-- ..((.............((((...(((.(((((....((((((((.........))))))))..))))))))....))))......(((.((((.........)))).))))).....-- ( -25.90) >consensus AAGCCAAACCAAAAUACCCCACUAAAUGCUAUAUCACUCGCACUUCAAAGUAAGAGGUGCGAAUUAUAGAUUGCAUUGGGUCUAUAGCUUUUGUAUUCUGAUCAUAGUAGCGCAUAAG__ ...........................(((((.....(((((((((.......)))))))))....((((.((((..((........))..)))).))))......)))))......... (-26.59 = -26.70 + 0.11)



| Location | 15,911,159 – 15,911,279 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -29.40 |
| Energy contribution | -30.07 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15911159 120 + 22224390 CCCAAUGCAAUCUAUAAUUCGCACCUCUUACUUUGAAGUGCGAGUGAUAUAGCAUUUAGUGGGGUAUUUUUGUUUGGCUUGGCUUUGAUUUGACUCUUGUUCGUGGUCACAGUAGCUAUA ((((....((((((((((((((((.((.......)).))))))))..))))).)))...))))...........(((((..(((.(((((.(((....)))...))))).)))))))).. ( -32.40) >DroSim_CAF1 6724 120 + 1 CCCAAUGCAAUCUAUAAUUCGCACCUCUUAGUUUGAAGUGCGAGUGAUAUAGCAUUUAGUGGGGUAUUUUGGUUUGGCUUGGCUUUGAUUUGACUCUUGUUCGUGGUCACAGUAGCUAUA ((((....((((((((((((((((.((.......)).))))))))..))))).)))...))))...........(((((..(((.(((((.(((....)))...))))).)))))))).. ( -32.40) >DroYak_CAF1 6915 120 + 1 CCCAAUGCAAUCUAUAAUUCGCACCUAUUACUUUGAGGUGCGAUUAAUAUAGCAUUUAAUGGGGUUUUUUGGUUUGGCUUGGCUUUGAUUUGACUCUUGUUCGUGGUCACAGUAGCUAUA ((((.((.((((((((..((((((((.........))))))))....))))).))))).))))...........(((((..(((.(((((.(((....)))...))))).)))))))).. ( -32.50) >consensus CCCAAUGCAAUCUAUAAUUCGCACCUCUUACUUUGAAGUGCGAGUGAUAUAGCAUUUAGUGGGGUAUUUUGGUUUGGCUUGGCUUUGAUUUGACUCUUGUUCGUGGUCACAGUAGCUAUA ((((.((.((((((((((((((((.((.......)).))))))))..))))).))))).))))...........(((((..(((.(((((.(((....)))...))))).)))))))).. (-29.40 = -30.07 + 0.67)



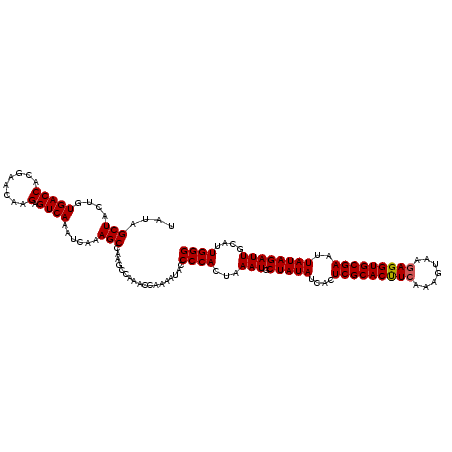
| Location | 15,911,159 – 15,911,279 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.46 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15911159 120 - 22224390 UAUAGCUACUGUGACCACGAACAAGAGUCAAAUCAAAGCCAAGCCAAACAAAAAUACCCCACUAAAUGCUAUAUCACUCGCACUUCAAAGUAAGAGGUGCGAAUUAUAGAUUGCAUUGGG ....(((....(((((........).))))......)))..................((((...(((.(((((....(((((((((.......)))))))))..))))))))....)))) ( -26.70) >DroSim_CAF1 6724 120 - 1 UAUAGCUACUGUGACCACGAACAAGAGUCAAAUCAAAGCCAAGCCAAACCAAAAUACCCCACUAAAUGCUAUAUCACUCGCACUUCAAACUAAGAGGUGCGAAUUAUAGAUUGCAUUGGG ....(((....(((((........).))))......)))..................((((...(((.(((((....(((((((((.......)))))))))..))))))))....)))) ( -26.70) >DroYak_CAF1 6915 120 - 1 UAUAGCUACUGUGACCACGAACAAGAGUCAAAUCAAAGCCAAGCCAAACCAAAAAACCCCAUUAAAUGCUAUAUUAAUCGCACCUCAAAGUAAUAGGUGCGAAUUAUAGAUUGCAUUGGG ....(((....(((((........).))))......)))..................((((...(((.(((((....((((((((.........))))))))..))))))))....)))) ( -25.70) >consensus UAUAGCUACUGUGACCACGAACAAGAGUCAAAUCAAAGCCAAGCCAAACCAAAAUACCCCACUAAAUGCUAUAUCACUCGCACUUCAAAGUAAGAGGUGCGAAUUAUAGAUUGCAUUGGG ....(((....(((((........).))))......)))..................((((...(((.(((((....(((((((((.......)))))))))..))))))))....)))) (-26.42 = -26.53 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:19 2006