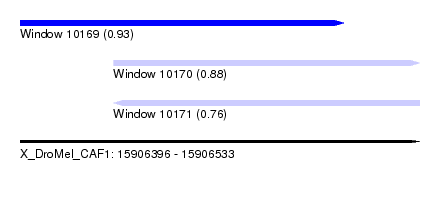
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,906,396 – 15,906,533 |
| Length | 137 |
| Max. P | 0.934653 |

| Location | 15,906,396 – 15,906,507 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.86 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -24.20 |
| Energy contribution | -23.44 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15906396 111 + 22224390 AUUAAC--------GUGUAGGAGUGUGAAUGCUAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCUUAAUGCUAAGUAUGACGGCUUGGAAAA .....(--------((.((((((((....(((((((((((....((((((((.....)))))))).......))))).))))-))..)))))))))))((((((......)))))).... ( -35.20) >DroSec_CAF1 1871 111 + 1 AUUAAC--------GUGUAGGAGUGUGAAUGCCAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCCUAAUGAUAACUAUGUCGUCUGGGAAAA ......--------...((((((((....(((((((((((....((((((((.....)))))))).......))))).))))-))..))))))))((((((....))))))......... ( -37.00) >DroSim_CAF1 1761 111 + 1 AUUAAC--------GUGUAGGAGUGUGAAUGCCAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCCUAAUGAUAACUAUGUCGGCUGGGAAAA ......--------...((((((((....(((((((((((....((((((((.....)))))))).......))))).))))-))..)))))))).(((((....))))).......... ( -36.30) >DroEre_CAF1 1842 112 + 1 AUUUAC--------GUGUAGGAGUGUGAACACCAGUGCUGGAUAGGGUAUGCGAUUUGUAUAACUACAAGGACGGUACCUAGAUAGGUAUAUUUAAUGAUAAGUAUGUCCGCCAGGAUAA ......--------......(.(((....)))).((.((((...((..((((..((((((....))))))....((((((....))))))............))))..)).)))).)).. ( -28.80) >DroYak_CAF1 1505 119 + 1 AUGUACCUAUCAAUGUGUAGGAGUGCGAAUACCAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUAG-CAGGUACUUUUCAAGAUAUGUUGGUCAGCCAAUAAGA .......((((....((((((.(..(((((....(((((......)))))...)))))..).)))))).(((.((((((...-..)))))).)))..))))((((((....))))))... ( -38.50) >consensus AUUAAC________GUGUAGGAGUGUGAAUGCCAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG_UAGGUACUCUUAAUGAUAAGUAUGUCGGCUAGGAAAA .................(((((((......(((.((.((.....((((((((.....))))))))...)).)))))((((....)))))))))))......................... (-24.20 = -23.44 + -0.76)

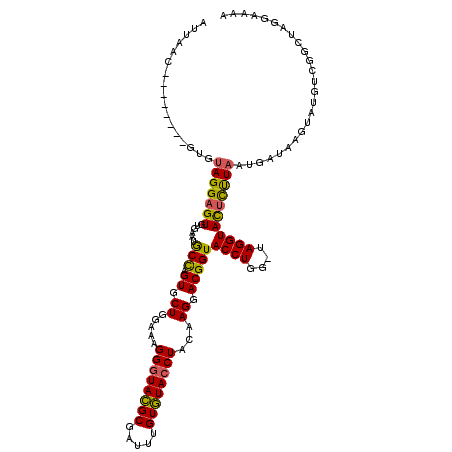
| Location | 15,906,428 – 15,906,533 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -19.26 |
| Energy contribution | -18.18 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15906428 105 + 22224390 GAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCUUAAUGCUAAGUAUGACGGCUUGGAAAAUGAGUAAGCAACAAC----UACUCUGCGUA---------- .......((((((....(.((((((((.(((......))).)-))))))).)......((((((......)))))).....(((((.........----)))))))))))---------- ( -30.30) >DroSec_CAF1 1903 115 + 1 GAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCCUAAUGAUAACUAUGUCGUCUGGGAAAAAUCGUAAGCAAUAUU----UACUCUGCACAUAUUUAGGGA ......((..(((((((..((((((((.(((......))).)-)))))))(((((((((((....)))))).))))).)))))))..))......----..(((((........))))). ( -34.80) >DroSim_CAF1 1793 115 + 1 GAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCCUAAUGAUAACUAUGUCGGCUGGGAAAAAUAGUAAGCAAUAUC----UACUCUGCACAUAUUUAGGGA ...((((((...(((((((((((((((.(((......))).)-)))))))(((((.(((((....)))))..)))))..........)))).)))----))))))............... ( -31.80) >DroEre_CAF1 1874 115 + 1 GAUAGGGUAUGCGAUUUGUAUAACUACAAGGACGGUACCUAGAUAGGUAUAUUUAAUGAUAAGUAUGUCCGCCAGGAUAAAUGGUAAGAAAUUAUUUUUUAUUGAGCUUAUAUUU----- ....((..((((..((((((....))))))....((((((....))))))............))))..))((...((((((..((((....))))..))))))..))........----- ( -25.30) >DroYak_CAF1 1545 110 + 1 GAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUAG-CAGGUACUUUUCAAGAUAUGUUGGUCAGCCAAUAAGAAUGGCAAGCAAUAAC----UACUAAGCUCAAAGCU----- ....((((((((.....))))))))....(((.((((((...-..)))))).)))......(((((.(..((((.......)))).).)))))..----.....(((.....)))----- ( -29.90) >consensus GAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG_UAGGUACUCUUAAUGAUAAGUAUGUCGGCUAGGAAAAAUAGUAAGCAAUAAC____UACUCUGCACAUAUUU_____ ....((((((((.....))))))))...((((..((((((....))))))))))................((((.......))))................................... (-19.26 = -18.18 + -1.08)



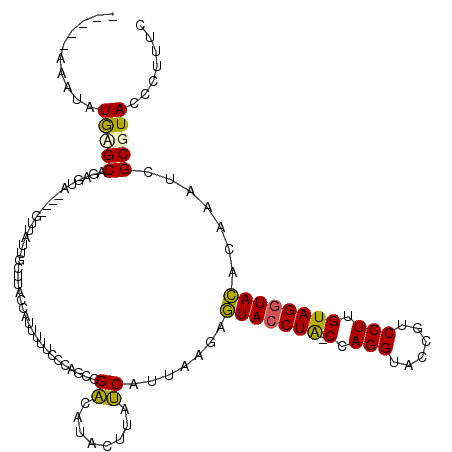
| Location | 15,906,428 – 15,906,533 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.10 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15906428 105 - 22224390 ----------UACGCAGAGUA----GUUGUUGCUUACUCAUUUUCCAAGCCGUCAUACUUAGCAUUAAGAGUACCUA-CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUC ----------(((((.(((((----((....)).)))))...............................(((((((-(.(((......))).)))))))).......)))))....... ( -29.50) >DroSec_CAF1 1903 115 - 1 UCCCUAAAUAUGUGCAGAGUA----AAUAUUGCUUACGAUUUUUCCCAGACGACAUAGUUAUCAUUAGGAGUACCUA-CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUC ...........((((.(((((----.....))))).((((((.(((..((..((...))..))....)))(((((((-(.(((......))).))))))))..)))))).))))...... ( -29.20) >DroSim_CAF1 1793 115 - 1 UCCCUAAAUAUGUGCAGAGUA----GAUAUUGCUUACUAUUUUUCCCAGCCGACAUAGUUAUCAUUAGGAGUACCUA-CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUC ..(((((.(((((((((((((----(.((.....))))))))).....))..))))).......))))).(((((((-(.(((......))).))))))))................... ( -25.10) >DroEre_CAF1 1874 115 - 1 -----AAAUAUAAGCUCAAUAAAAAAUAAUUUCUUACCAUUUAUCCUGGCGGACAUACUUAUCAUUAAAUAUACCUAUCUAGGUACCGUCCUUGUAGUUAUACAAAUCGCAUACCCUAUC -----........((................................(((((......(((....)))...(((((....)))))))))).(((((....)))))...)).......... ( -14.30) >DroYak_CAF1 1545 110 - 1 -----AGCUUUGAGCUUAGUA----GUUAUUGCUUGCCAUUCUUAUUGGCUGACCAACAUAUCUUGAAAAGUACCUG-CUAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUC -----.((...((((......----......))))((((.......))))....................(((((((-(.(((......))).)))))))).......)).......... ( -26.40) >consensus _____AAAUAUGAGCAGAGUA____GUUAUUGCUUACCAUUUUUCCCAGCCGACAUACUUAUCAUUAAGAGUACCUA_CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUC ..........(((((....................................((........)).......(((((((.(.(((......))).)))))))).......)))))....... (-13.62 = -13.10 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:11 2006