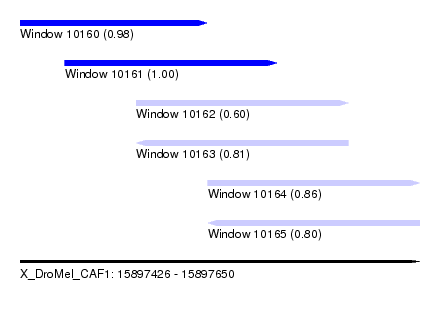
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,897,426 – 15,897,650 |
| Length | 224 |
| Max. P | 0.998254 |

| Location | 15,897,426 – 15,897,531 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -19.95 |
| Energy contribution | -21.23 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15897426 105 + 22224390 --------------AAC-CAACUUUCGCUACCAGCUCAUGAAUUUCCCAACUUCAAUAAAAUCCCUGGUGAAAGCUUUGGGACAGACCUUUCUUUCCCCAACUAGGGGAGAUGUCCCGAG --------------...-...(((((((((........((((.........))))..........)))))))))..((((((((((....))(((((((.....))))))))))))))). ( -29.17) >DroSec_CAF1 4903 119 + 1 GGAAGCUAAAAAGCAAC-CAACUUUCGCUACCAGCCCAUGAAUUUCCCAACUUUAAUAAACUGCCUGGUGAAAGCUUUGGGACAGACUUCUCUUUCCCCAACUAGGGGAGAUGUCCUGAG ((..(((....)))..)-)..(((((((((.(((..........................)))..)))))))))..(..(((((((....))(((((((.....))))))))))))..). ( -32.27) >DroSim_CAF1 4564 117 + 1 --AAGCUGAAAAGCAAC-CAACUUUCGCUACCAGCCCAUGAAUUUCCCAACUUUAAUAAAAUGCCUGGUGAAAGCUUUGGGACGAACUUCUCUUUCCCCAACUAGGGGAGAUGUCCUGAG --..(((....)))...-...(((((...(((((..(((.....................))).))))))))))..(..((((((.....))(((((((.....))))))).))))..). ( -30.80) >DroEre_CAF1 20867 105 + 1 ----------AAGAAACCCAACUUG----UCCAAGCAAUCAAUUUCCCAACUUCAAUAAAACCCUUCGUGAAAGCUUUAGGACAAU-GAAAGUUUCCCCAACCAGGGGAGAUGUCCUGAG ----------............(((----(((((((..(((...........................)))..))))..)))))).-....((((((((.....))))))))........ ( -21.43) >DroYak_CAF1 5235 105 + 1 ----------AAGAAACCAAACUUU----UCCAGGCAAUGAAUUUCCCAACUUUAAUAAAAUCCUAAGUAAAAGCUUUGGGACAAG-AAAAGUUUCCCCAAUAAGGGGAGAUGUCCUGAG ----------...............----..(((((........((((((((((.((..........)).))))..))))))....-....((((((((.....))))))))).)))).. ( -24.10) >consensus __________AAGAAAC_CAACUUUCGCUACCAGCCCAUGAAUUUCCCAACUUUAAUAAAAUCCCUGGUGAAAGCUUUGGGACAAACUUAUCUUUCCCCAACUAGGGGAGAUGUCCUGAG .....................(((((((((...................................)))))))))..((((((((........(((((((.....))))))))))))))). (-19.95 = -21.23 + 1.28)


| Location | 15,897,451 – 15,897,570 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.20 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.72 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15897451 119 + 22224390 AAUUUCCCAACUUCAAUAAAAUCCCUGGUGAAAGCUUUGGGACAGACCUUUCUUUCCCCAACUAGGGGAGAUGUCCCGAGCAGUGUUGAGUAUCCGUAUGGACUCAACAGAAAUCCCCU- ..(((((((................))).))))((((.((((((((....))(((((((.....)))))))))))))))))..(((((((..(((....))))))))))..........- ( -37.69) >DroSec_CAF1 4942 120 + 1 AAUUUCCCAACUUUAAUAAACUGCCUGGUGAAAGCUUUGGGACAGACUUCUCUUUCCCCAACUAGGGGAGAUGUCCUGAGCAGUGUUGAGUACCCGUAUGGACUCAACAGAAAUCCCCUU .(((((.............(((((..(((....)))(..(((((((....))(((((((.....))))))))))))..))))))(((((((.((.....))))))))).)))))...... ( -39.30) >DroSim_CAF1 4601 120 + 1 AAUUUCCCAACUUUAAUAAAAUGCCUGGUGAAAGCUUUGGGACGAACUUCUCUUUCCCCAACUAGGGGAGAUGUCCUGAGCAGUGUUGAGUACCCGUAUGGACUCAACAGAAAUCCCCUU ..(((((((................))).))))((.(..((((((.....))(((((((.....))))))).))))..)))..((((((((.((.....))))))))))........... ( -35.99) >DroEre_CAF1 20893 117 + 1 AAUUUCCCAACUUCAAUAAAACCCUUCGUGAAAGCUUUAGGACAAU-GAAAGUUUCCCCAACCAGGGGAGAUGUCCUGAGUAGUGGUGAGUA-CAUUAAGGACUCAACAGAAAUCCCCU- .((((((((.((.(.............((....)).((((((((..-.....(((((((.....)))))))))))))))).)))))(((((.-(.....).)))))...))))).....- ( -29.31) >DroYak_CAF1 5261 118 + 1 AAUUUCCCAACUUUAAUAAAAUCCUAAGUAAAAGCUUUGGGACAAG-AAAAGUUUCCCCAAUAAGGGGAGAUGUCCUGAGCAGUGGUGAGUAGCAUAAAGGACUCAACAGAAAUCCCCU- .((((((((((((............))))....((.(..(((((..-.....(((((((.....))))))))))))..)))..)))(((((..(.....).)))))...))))).....- ( -28.81) >consensus AAUUUCCCAACUUUAAUAAAAUCCCUGGUGAAAGCUUUGGGACAAACUUAUCUUUCCCCAACUAGGGGAGAUGUCCUGAGCAGUGUUGAGUACCCGUAUGGACUCAACAGAAAUCCCCU_ .(((((...........................((((.((((((........(((((((.....)))))))))))))))))..((((((((.((.....)))))))))))))))...... (-30.64 = -30.72 + 0.08)

| Location | 15,897,491 – 15,897,610 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -37.09 |
| Consensus MFE | -30.06 |
| Energy contribution | -30.66 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15897491 119 + 22224390 GACAGACCUUUCUUUCCCCAACUAGGGGAGAUGUCCCGAGCAGUGUUGAGUAUCCGUAUGGACUCAACAGAAAUCCCCU-UUGAGUGACUCACUCACUGGAUUCGAGUGUGCAGACAAGC ............(((((((.....)))))))((((....(((.(((((((..(((....))))))))))(((.(((...-.((((((...))))))..))))))...)))...))))... ( -35.70) >DroSec_CAF1 4982 120 + 1 GACAGACUUCUCUUUCCCCAACUAGGGGAGAUGUCCUGAGCAGUGUUGAGUACCCGUAUGGACUCAACAGAAAUCCCCUUUUGAGUGACUCACUCACGGGAUUCGAGUGUGCAGACAAGC ............(((((((.....)))))))((((.((.(((.((((((((.((.....))))))))))(((.((((....((((((...)))))).)))))))...))).))))))... ( -41.70) >DroSim_CAF1 4641 120 + 1 GACGAACUUCUCUUUCCCCAACUAGGGGAGAUGUCCUGAGCAGUGUUGAGUACCCGUAUGGACUCAACAGAAAUCCCCUUUUGAGUGACUCACUCACGGGAUUCGAGUGUGCAGACAAGC ............(((((((.....)))))))((((.((.(((.((((((((.((.....))))))))))(((.((((....((((((...)))))).)))))))...))).))))))... ( -41.70) >DroEre_CAF1 20933 117 + 1 GACAAU-GAAAGUUUCCCCAACCAGGGGAGAUGUCCUGAGUAGUGGUGAGUA-CAUUAAGGACUCAACAGAAAUCCCCU-CAGAGUGACUCACUCACAGGAUUUGAGUGUGCAGACAAGC ......-....((((((((.....))))))))(((.(((((..(((((....-)))))...)))))((..((((((...-..(((((...)))))...))))))..)).....))).... ( -32.60) >DroYak_CAF1 5301 118 + 1 GACAAG-AAAAGUUUCCCCAAUAAGGGGAGAUGUCCUGAGCAGUGGUGAGUAGCAUAAAGGACUCAACAGAAAUCCCCU-CCGAGUGGCUCACUCACAGGAUUUGAGUGUGCAGACCAGC ..((.(-(...((((((((.....)))))))).)).)).((.(((((((((.((....(((...............)))-....)).)))))).))).((.((((......)))))).)) ( -33.76) >consensus GACAAACUUAUCUUUCCCCAACUAGGGGAGAUGUCCUGAGCAGUGUUGAGUACCCGUAUGGACUCAACAGAAAUCCCCU_UUGAGUGACUCACUCACAGGAUUCGAGUGUGCAGACAAGC .(((........(((((((.....))))))).((((((.....((((((((.((.....)))))))))).............(((((...))))).)))))).....))).......... (-30.06 = -30.66 + 0.60)

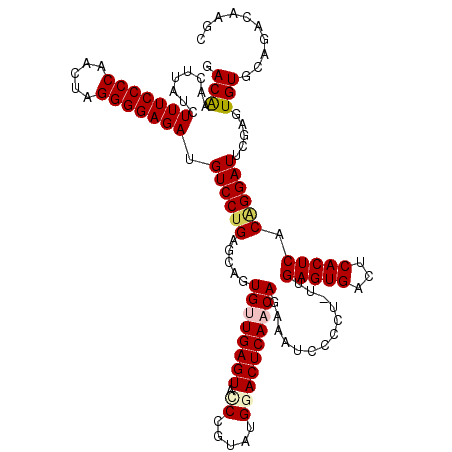
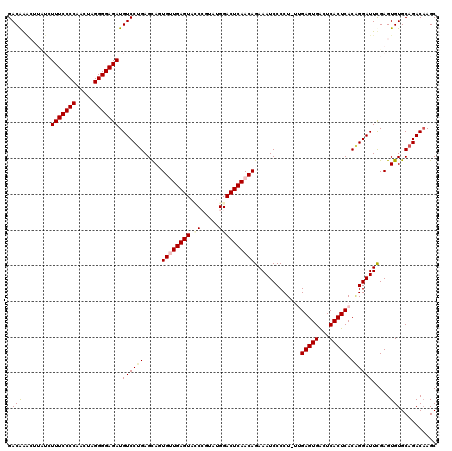
| Location | 15,897,491 – 15,897,610 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -31.36 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.813051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15897491 119 - 22224390 GCUUGUCUGCACACUCGAAUCCAGUGAGUGAGUCACUCAA-AGGGGAUUUCUGUUGAGUCCAUACGGAUACUCAACACUGCUCGGGACAUCUCCCCUAGUUGGGGAAAGAAAGGUCUGUC ((((.(((.....(((((((((..((((((...)))))).-...))))...((((((((((....)))..)))))))....))))).....(((((.....))))).))).))))..... ( -39.00) >DroSec_CAF1 4982 120 - 1 GCUUGUCUGCACACUCGAAUCCCGUGAGUGAGUCACUCAAAAGGGGAUUUCUGUUGAGUCCAUACGGGUACUCAACACUGCUCAGGACAUCUCCCCUAGUUGGGGAAAGAGAAGUCUGUC ((((.((((((.....(((((((.((((((...))))))....))))))).((((((((((.....)).)))))))).)))..........(((((.....))))).))).))))..... ( -43.50) >DroSim_CAF1 4641 120 - 1 GCUUGUCUGCACACUCGAAUCCCGUGAGUGAGUCACUCAAAAGGGGAUUUCUGUUGAGUCCAUACGGGUACUCAACACUGCUCAGGACAUCUCCCCUAGUUGGGGAAAGAGAAGUUCGUC ((((.((((((.....(((((((.((((((...))))))....))))))).((((((((((.....)).)))))))).)))..........(((((.....))))).))).))))..... ( -43.50) >DroEre_CAF1 20933 117 - 1 GCUUGUCUGCACACUCAAAUCCUGUGAGUGAGUCACUCUG-AGGGGAUUUCUGUUGAGUCCUUAAUG-UACUCACCACUACUCAGGACAUCUCCCCUGGUUGGGGAAACUUUC-AUUGUC ((......)).........(((((((.((((((.((..((-((((...(((....)))))))))..)-)))))))))).....))))....(((((.....))))).......-...... ( -32.30) >DroYak_CAF1 5301 118 - 1 GCUGGUCUGCACACUCAAAUCCUGUGAGUGAGCCACUCGG-AGGGGAUUUCUGUUGAGUCCUUUAUGCUACUCACCACUGCUCAGGACAUCUCCCCUUAUUGGGGAAACUUUU-CUUGUC ((((((..(((.........(((.((((((...)))))).-)))((((((.....))))))....))).....))))..)).(((((....(((((.....)))))......)-)))).. ( -34.40) >consensus GCUUGUCUGCACACUCGAAUCCCGUGAGUGAGUCACUCAA_AGGGGAUUUCUGUUGAGUCCAUACGGGUACUCAACACUGCUCAGGACAUCUCCCCUAGUUGGGGAAAGAGAAGUUUGUC ....(((((((.....(((((((.((((((...))))))....))))))).((((((((((....))..)))))))).)))...))))...(((((.....))))).............. (-31.36 = -32.24 + 0.88)

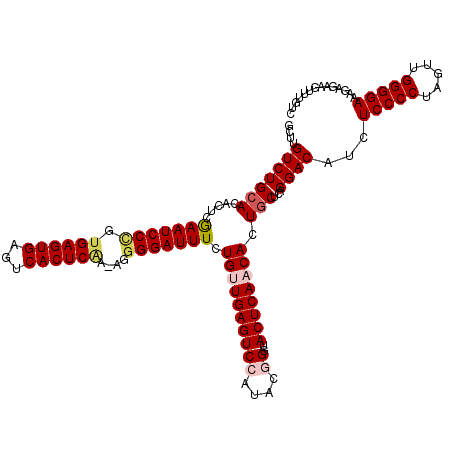
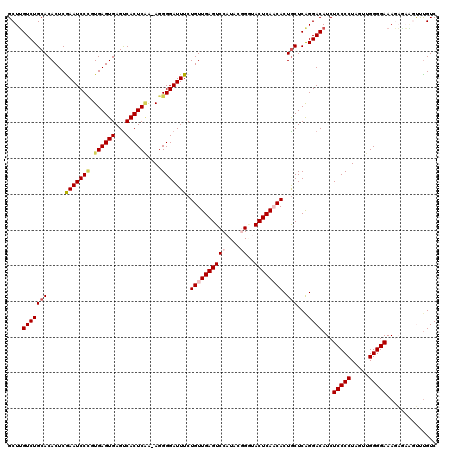
| Location | 15,897,531 – 15,897,650 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.31 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -32.02 |
| Energy contribution | -32.78 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15897531 119 + 22224390 CAGUGUUGAGUAUCCGUAUGGACUCAACAGAAAUCCCCU-UUGAGUGACUCACUCACUGGAUUCGAGUGUGCAGACAAGCUUUCGCUCAUUUGUGUUCGGCUUUUCAUUUGAACGCUUGG ...(((((((..(((....))))))))))..(((((...-.((((((...))))))..)))))(((((((.((((.(((((..(((......)))...)))))....)))).))))))). ( -36.50) >DroSec_CAF1 5022 120 + 1 CAGUGUUGAGUACCCGUAUGGACUCAACAGAAAUCCCCUUUUGAGUGACUCACUCACGGGAUUCGAGUGUGCAGACAAGCUUUCGCUCAUUUGUGCUCGGCUUUUCAUUUGGACGCUUGG ...((((((((.((.....))))))))))..((((((....((((((...)))))).))))))(((((((.((((.(((((..(((......)))...)))))....)))).))))))). ( -42.30) >DroSim_CAF1 4681 120 + 1 CAGUGUUGAGUACCCGUAUGGACUCAACAGAAAUCCCCUUUUGAGUGACUCACUCACGGGAUUCGAGUGUGCAGACAAGCUUUCGCUCAUUUGUGCUCGGCUUUUCAUUUGGACGCUUGG ...((((((((.((.....))))))))))..((((((....((((((...)))))).))))))(((((((.((((.(((((..(((......)))...)))))....)))).))))))). ( -42.30) >DroEre_CAF1 20972 118 + 1 UAGUGGUGAGUA-CAUUAAGGACUCAACAGAAAUCCCCU-CAGAGUGACUCACUCACAGGAUUUGAGUGUGCAGACAAGCUUUCGCUCACUUGCGCUCGGCUUUUCAUUUGGACGCUUGG ..(((((((((.-((((.(((...............)))-...)))))))))).))).....(..(((((.((((.(((((..(((......)))...)))))....)))).)))))..) ( -35.36) >DroYak_CAF1 5340 119 + 1 CAGUGGUGAGUAGCAUAAAGGACUCAACAGAAAUCCCCU-CCGAGUGGCUCACUCACAGGAUUUGAGUGUGCAGACCAGCUUUCACUCACUUGUGCUCGGCUUUUCAUUUGGACGCUUGG .((((.(((((..(.....).)))))............(-((((((((((....((((((...((((((.((......))...))))))))))))...)))....))))))))))))... ( -32.30) >consensus CAGUGUUGAGUACCCGUAUGGACUCAACAGAAAUCCCCU_UUGAGUGACUCACUCACAGGAUUCGAGUGUGCAGACAAGCUUUCGCUCAUUUGUGCUCGGCUUUUCAUUUGGACGCUUGG ...((((((((.((.....))))))))))..(((((.....((((((...))))))..)))))(((((((.((((.(((((..(((......)))...)))))....)))).))))))). (-32.02 = -32.78 + 0.76)


| Location | 15,897,531 – 15,897,650 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.31 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -28.24 |
| Energy contribution | -28.56 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15897531 119 - 22224390 CCAAGCGUUCAAAUGAAAAGCCGAACACAAAUGAGCGAAAGCUUGUCUGCACACUCGAAUCCAGUGAGUGAGUCACUCAA-AGGGGAUUUCUGUUGAGUCCAUACGGAUACUCAACACUG ....((((((...(....)...))))....(..(((....)))..)..))......((((((..((((((...)))))).-...)))))).((((((((((....)))..)))))))... ( -35.90) >DroSec_CAF1 5022 120 - 1 CCAAGCGUCCAAAUGAAAAGCCGAGCACAAAUGAGCGAAAGCUUGUCUGCACACUCGAAUCCCGUGAGUGAGUCACUCAAAAGGGGAUUUCUGUUGAGUCCAUACGGGUACUCAACACUG ....((.((.....))...))((((..((.(..(((....)))..).))....))))((((((.((((((...))))))....))))))..((((((((((.....)).))))))))... ( -39.20) >DroSim_CAF1 4681 120 - 1 CCAAGCGUCCAAAUGAAAAGCCGAGCACAAAUGAGCGAAAGCUUGUCUGCACACUCGAAUCCCGUGAGUGAGUCACUCAAAAGGGGAUUUCUGUUGAGUCCAUACGGGUACUCAACACUG ....((.((.....))...))((((..((.(..(((....)))..).))....))))((((((.((((((...))))))....))))))..((((((((((.....)).))))))))... ( -39.20) >DroEre_CAF1 20972 118 - 1 CCAAGCGUCCAAAUGAAAAGCCGAGCGCAAGUGAGCGAAAGCUUGUCUGCACACUCAAAUCCUGUGAGUGAGUCACUCUG-AGGGGAUUUCUGUUGAGUCCUUAAUG-UACUCACCACUA ....((.((.....))...)).(((.(((.(..(((....)))..).)))...))).......(((.((((((.((..((-((((...(((....)))))))))..)-)))))))))).. ( -35.10) >DroYak_CAF1 5340 119 - 1 CCAAGCGUCCAAAUGAAAAGCCGAGCACAAGUGAGUGAAAGCUGGUCUGCACACUCAAAUCCUGUGAGUGAGCCACUCGG-AGGGGAUUUCUGUUGAGUCCUUUAUGCUACUCACCACUG ...(((((............(((((....(((.(((....))).).))((.((((((.......)))))).))..)))))-(((((((((.....))))))))))))))........... ( -31.60) >consensus CCAAGCGUCCAAAUGAAAAGCCGAGCACAAAUGAGCGAAAGCUUGUCUGCACACUCGAAUCCCGUGAGUGAGUCACUCAA_AGGGGAUUUCUGUUGAGUCCAUACGGGUACUCAACACUG ....((.((.....))...)).(((..((.(..(((....)))..).))....)))(((((((.((((((...))))))....))))))).((((((((((....))..))))))))... (-28.24 = -28.56 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:06 2006