
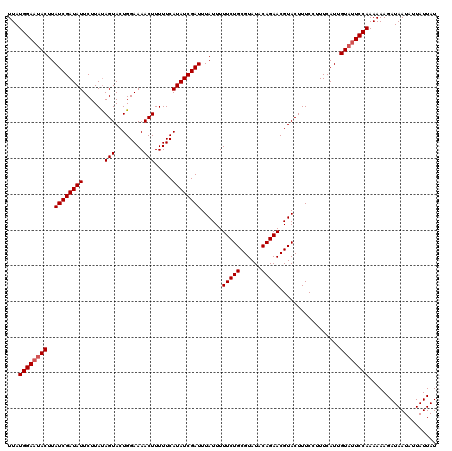
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,826,361 – 15,826,677 |
| Length | 316 |
| Max. P | 0.994655 |

| Location | 15,826,361 – 15,826,481 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -20.63 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
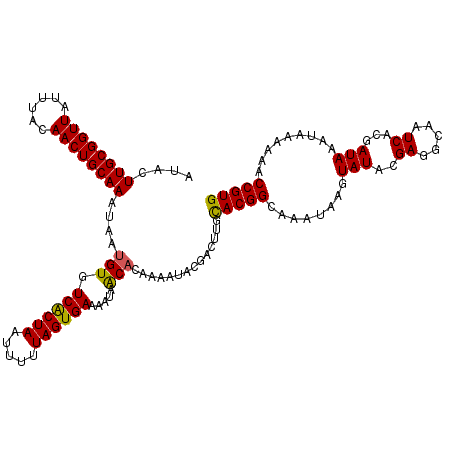
>X_DroMel_CAF1 15826361 120 + 22224390 UUAUGGAAUACUUAUCGAUAUUCUUACAGUACUGGAAAACUUUUUCAUAUCGAUUUAUUUUUCUGCGUAUACAGAACGUACUUUCGUUUCAUUGUUGUCCAAAAAAGAUAAUAAUAUUAU ..(((((((((..((((((((.....((....))((((....))))))))))))......(((((......))))).))).))))))...(((((((((.......)))))))))..... ( -20.10) >DroSec_CAF1 5229 120 + 1 UUAUGGAAUACUUAUCGAUAUUCUUAUAGUACUGGAAAACUUUUUCAUAUCGAUUUAUUUUUCUGCGUAUACAGAACGUACUUUCCUUUCAUUGUAUUCCAAAAAAGAUAAUAUUAUUAU ...((((((((..((((((((......(((........))).....))))))))......(((((......))))).................))))))))................... ( -20.90) >DroSim_CAF1 5018 120 + 1 UUAUGGAAUACUUAUCGAUAUUCUUAUAGUACUGGAAAACUUUUUCAUAUCGAUUUAUUUUUCUGCGUAUACAGAACGUACUUUCCUUUCAUUGUAUUCCAAAAAAGAUAAUAUUAUUAU ...((((((((..((((((((......(((........))).....))))))))......(((((......))))).................))))))))................... ( -20.90) >consensus UUAUGGAAUACUUAUCGAUAUUCUUAUAGUACUGGAAAACUUUUUCAUAUCGAUUUAUUUUUCUGCGUAUACAGAACGUACUUUCCUUUCAUUGUAUUCCAAAAAAGAUAAUAUUAUUAU ...((((((((..((((((((......(((........))).....))))))))......(((((......))))).................))))))))................... (-18.90 = -19.57 + 0.67)


| Location | 15,826,361 – 15,826,481 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15826361 120 - 22224390 AUAAUAUUAUUAUCUUUUUUGGACAACAAUGAAACGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAAGUUUUCCAGUACUGUAAGAAUAUCGAUAAGUAUUCCAUAA ...................((((..((......((....))...((((((....))))))......((((((((.....(((........)))......))))))))..))..))))... ( -21.10) >DroSec_CAF1 5229 120 - 1 AUAAUAAUAUUAUCUUUUUUGGAAUACAAUGAAAGGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAAGUUUUCCAGUACUAUAAGAAUAUCGAUAAGUAUUCCAUAA ...................((((((((.................((((((....))))))......((((((((.....(((........)))......))))))))..))))))))... ( -24.50) >DroSim_CAF1 5018 120 - 1 AUAAUAAUAUUAUCUUUUUUGGAAUACAAUGAAAGGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAAGUUUUCCAGUACUAUAAGAAUAUCGAUAAGUAUUCCAUAA ...................((((((((.................((((((....))))))......((((((((.....(((........)))......))))))))..))))))))... ( -24.50) >consensus AUAAUAAUAUUAUCUUUUUUGGAAUACAAUGAAAGGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAAGUUUUCCAGUACUAUAAGAAUAUCGAUAAGUAUUCCAUAA ...................((((((((.................((((((....))))))......((((((((.....(((........)))......))))))))..))))))))... (-23.10 = -23.77 + 0.67)


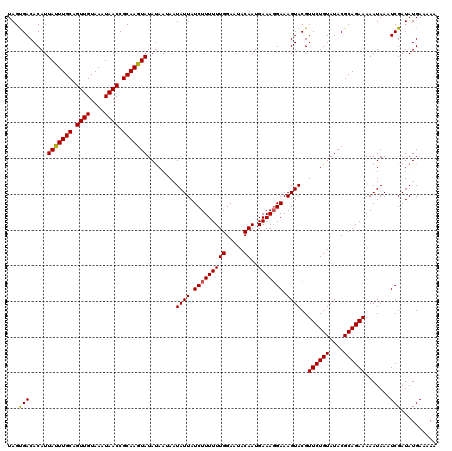
| Location | 15,826,401 – 15,826,521 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.48 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15826401 120 - 22224390 UAGUGACACAUUACUUGCAGUUGUAAAUAACCGCAAGUAUAUAAUAUUAUUAUCUUUUUUGGACAACAAUGAAACGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAA (((((......(((((((.((((....)))).))))))).....))))).(((((((.(((.....))).)))((....))...((((((....)))))).........))))....... ( -20.80) >DroSec_CAF1 5269 120 - 1 UAGUGACACAUUAUUUGCAGUUGUAAAUAACCGCAAGUAUAUAAUAAUAUUAUCUUUUUUGGAAUACAAUGAAAGGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAA ...........(((((((.((((....)))).)))))))........((((.(((((((((.....))..))))))).))))..((((((....)))))).................... ( -18.80) >DroSim_CAF1 5058 120 - 1 UAGUGACACAUUAUUUGCAGUUGUAAAUAACCGCAAGUAUAUAAUAAUAUUAUCUUUUUUGGAAUACAAUGAAAGGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAA ...........(((((((.((((....)))).)))))))........((((.(((((((((.....))..))))))).))))..((((((....)))))).................... ( -18.80) >DroEre_CAF1 5775 120 - 1 UAGCGACACAUUAUUUGCAGUUGUAAAUAACCGCAAGUAUAUAAAAAUAUUAUCUUUUUUGGAAUACAAUGAAAGGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAA ...(((.....(((((((.((((....)))).)))))))........((((.(((((((((.....))..))))))).))))..((((((....)))))).......))).......... ( -20.20) >DroYak_CAF1 5071 120 - 1 UAGCGACACAUUACUUGCAGUUGUAAAUAACCGCAAGUAUACAAAAAUAUUAUCUUUUUUGGAAUACAAUGAAAGGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAA ..(((......(((((((.((((....)))).)))))))((((((..((((.(((((((((.....))..))))))).))))..)).))))..)))........................ ( -23.00) >consensus UAGUGACACAUUAUUUGCAGUUGUAAAUAACCGCAAGUAUAUAAUAAUAUUAUCUUUUUUGGAAUACAAUGAAAGGAAAGUACGUUCUGUAUACGCAGAAAAAUAAAUCGAUAUGAAAAA ...(((.....(((((((.((((....)))).)))))))........((((.(((((((((.....))..))))))).))))..((((((....)))))).......))).......... (-19.76 = -19.48 + -0.28)


| Location | 15,826,481 – 15,826,601 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.39 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15826481 120 + 22224390 AUACUUGCGGUUAUUUACAACUGCAAGUAAUGUGUCACUAAUUUUUAGUGAAAAUAACACAAAAUACGACUUGUACGGCAAAUAAGUAUACGAGGCAAUCACGAUAAAUAAAAAACCGUG .(((((((((((......))))))))))).((((((((((.....))))))......))))........((((((..((......)).)))))).....((((.............)))) ( -28.12) >DroSec_CAF1 5349 120 + 1 AUACUUGCGGUUAUUUACAACUGCAAAUAAUGUGUCACUAAUUUUUAGUGAAAAUAGCACAAAAUACGACUUGCACGGCAAAUAAGUAUACGAGGCAAUCACGAUAAAUAAAAAACCGUG ....((((((((......))))))))....((((((((((.....)))))).....((.....((((...((((...))))....)))).....))...))))................. ( -22.20) >DroSim_CAF1 5138 120 + 1 AUACUUGCGGUUAUUUACAACUGCAAAUAAUGUGUCACUAAUUUUUAGUGAAAAUAGCACGAAAUACGACUUGCACGGCAAAUAAGUAUACGAGGCAAUCACGAUAAAUAAAAAACCGUG ....((((((((......))))))))....((((((((((.....)))))).....((.((..((((...((((...))))....)))).))..))...))))................. ( -22.40) >DroEre_CAF1 5855 119 + 1 AUACUUGCGGUUAUUUACAACUGCAAAUAAUGUGUCGCUAAUUUGUAGUGAAAAUAACCCAAAAUACGACUUGCACGGCAUAUAAAUAUAAGAGGC-AUCACUAUAAAUAAAAAACCGUG ....(((.((((((((...(((((((((...(.....)..)))))))))..)))))))))))...........(((((..............((..-....))............))))) ( -23.17) >DroYak_CAF1 5151 120 + 1 AUACUUGCGGUUAUUUACAACUGCAAGUAAUGUGUCGCUAAACUUUAGUGAAAAUAACAGAAAAUACGACUUGCACGGCAAAUAAAUAUAAGAGGCAAUCACGAUAAAUAAAAAACCGUG .(((((((((((......))))))))))).(((.((((((.....)))))).....)))..............(((((........(((..((.....))...))).........))))) ( -24.53) >consensus AUACUUGCGGUUAUUUACAACUGCAAAUAAUGUGUCACUAAUUUUUAGUGAAAAUAACACAAAAUACGACUUGCACGGCAAAUAAGUAUACGAGGCAAUCACGAUAAAUAAAAAACCGUG ....((((((((......))))))))....(((.((((((.....)))))).....)))..............(((((........(((..((.....))...))).........))))) (-18.83 = -18.39 + -0.44)


| Location | 15,826,481 – 15,826,601 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.76 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15826481 120 - 22224390 CACGGUUUUUUAUUUAUCGUGAUUGCCUCGUAUACUUAUUUGCCGUACAAGUCGUAUUUUGUGUUAUUUUCACUAAAAAUUAGUGACACAUUACUUGCAGUUGUAAAUAACCGCAAGUAU .(((((....((..(((((.(.....).)).)))..))...))))).............(((((((((.............))))))))).(((((((.((((....)))).))))))). ( -25.62) >DroSec_CAF1 5349 120 - 1 CACGGUUUUUUAUUUAUCGUGAUUGCCUCGUAUACUUAUUUGCCGUGCAAGUCGUAUUUUGUGCUAUUUUCACUAAAAAUUAGUGACACAUUAUUUGCAGUUGUAAAUAACCGCAAGUAU ((((((.........))))))...............(((((((.(..((((......))))..).....((((((.....))))))....((((((((....))))))))..))))))). ( -27.00) >DroSim_CAF1 5138 120 - 1 CACGGUUUUUUAUUUAUCGUGAUUGCCUCGUAUACUUAUUUGCCGUGCAAGUCGUAUUUCGUGCUAUUUUCACUAAAAAUUAGUGACACAUUAUUUGCAGUUGUAAAUAACCGCAAGUAU ((((((.........))))))...((..((.((((..((((((...)))))).))))..)).)).....((((((.....)))))).....(((((((.((((....)))).))))))). ( -26.60) >DroEre_CAF1 5855 119 - 1 CACGGUUUUUUAUUUAUAGUGAU-GCCUCUUAUAUUUAUAUGCCGUGCAAGUCGUAUUUUGGGUUAUUUUCACUACAAAUUAGCGACACAUUAUUUGCAGUUGUAAAUAACCGCAAGUAU ((((((....((..(((((.((.-...)))))))..))...))))))...(((((..(((((((.......))).))))...)))))....(((((((.((((....)))).))))))). ( -27.50) >DroYak_CAF1 5151 120 - 1 CACGGUUUUUUAUUUAUCGUGAUUGCCUCUUAUAUUUAUUUGCCGUGCAAGUCGUAUUUUCUGUUAUUUUCACUAAAGUUUAGCGACACAUUACUUGCAGUUGUAAAUAACCGCAAGUAU ((((((....((..(((...((.....))..)))..))...))))))...((((((((((..((.......)).)))))...)))))....(((((((.((((....)))).))))))). ( -24.10) >consensus CACGGUUUUUUAUUUAUCGUGAUUGCCUCGUAUACUUAUUUGCCGUGCAAGUCGUAUUUUGUGUUAUUUUCACUAAAAAUUAGUGACACAUUAUUUGCAGUUGUAAAUAACCGCAAGUAU ((((((....((..(((((.(.....).)).)))..))...))))))......................((((((.....)))))).....(((((((.((((....)))).))))))). (-20.44 = -20.76 + 0.32)


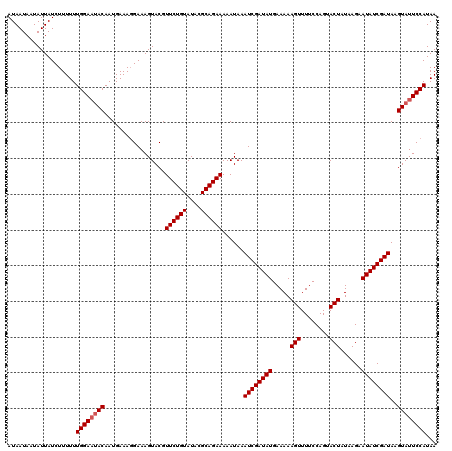
| Location | 15,826,561 – 15,826,677 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -23.84 |
| Energy contribution | -23.56 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15826561 116 - 22224390 AAGGUGUAUGUACAUAUCCACACAUCGAUAU----AUGUGCAUGUAAGUAUUGAGAAGCGUCCACUUGAAAUUCCAUACUCACGGUUUUUUAUUUAUCGUGAUUGCCUCGUAUACUUAUU ......(((((((((((............))----)))))))))(((((((((((..(((..(((.(((((..((........))..)))))......)))..))))))).))))))).. ( -25.90) >DroSec_CAF1 5429 116 - 1 AAGGUGUAUGUACAUAUCCACACAUCGAUAU----AUGUGCAUGUAAGUAUUGAGAAGCGUCCACUUGAAAUUCCAUACUCACGGUUUUUUAUUUAUCGUGAUUGCCUCGUAUACUUAUU ......(((((((((((............))----)))))))))(((((((((((..(((..(((.(((((..((........))..)))))......)))..))))))).))))))).. ( -25.90) >DroSim_CAF1 5218 116 - 1 AAGGUGUAUGUACAUAUCCACACAUCGAUAU----AUGUGCAUGUAAGUAUUGAGAAGCGUCCACUUGAAAUUCCAUACUCACGGUUUUUUAUUUAUCGUGAUUGCCUCGUAUACUUAUU ......(((((((((((............))----)))))))))(((((((((((..(((..(((.(((((..((........))..)))))......)))..))))))).))))))).. ( -25.90) >DroEre_CAF1 5935 119 - 1 AAGGUGUAUGUACAUAUCCACACAUCGAUAUACAUAUAUGCAUGUAAGUAUUGAGAAGCGUCCACUUGAAAUUCCAUACUCACGGUUUUUUAUUUAUAGUGAU-GCCUCUUAUAUUUAUA ...(((((((((.(((((........)))))...)))))))))((((((((.((((.(((((.((((((((..((........))..))))).....))))))-)).)))))))))))). ( -29.30) >DroYak_CAF1 5231 116 - 1 AAGGUGUAUGUACAUAUCCACACAUCGAUAU----AUAUGCAUGUAAGUAUUGAGAAGCGUCCACUUGAAAUUCCAUACUCACGGUUUUUUAUUUAUCGUGAUUGCCUCUUAUAUUUAUU ...(((((((((..((((........)))))----))))))))((((((((.((((.(((..(((.(((((..((........))..)))))......)))..))).)))))))))))). ( -25.40) >consensus AAGGUGUAUGUACAUAUCCACACAUCGAUAU____AUGUGCAUGUAAGUAUUGAGAAGCGUCCACUUGAAAUUCCAUACUCACGGUUUUUUAUUUAUCGUGAUUGCCUCGUAUACUUAUU ...((((((((..(((((........)))))....))))))))((((((((...((.(((..(((.(((((..((........))..)))))......)))..))).))..)))))))). (-23.84 = -23.56 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:42 2006