
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,816,485 – 15,816,602 |
| Length | 117 |
| Max. P | 0.999338 |

| Location | 15,816,485 – 15,816,602 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -30.84 |
| Energy contribution | -30.64 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
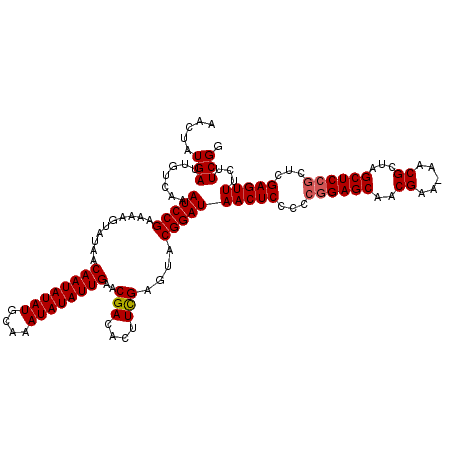
>X_DroMel_CAF1 15816485 117 + 22224390 CCGAAGAAACUCGAGCGGAGCUAGCGUU-UUCGUUGCUCCGG--GAGUUAUCCGUACUCGAAGUGUCGUUCAAUAUAUUUGCAUAUAUUGUUAUACUUUUCGGAUUUGACAUUCAUAGUU (((((((...(((((((((((.((((..-..))))))))))(--((....)))...))))).((((....((((((((....))))))))..)))))))))))................. ( -37.90) >DroSec_CAF1 16808 119 + 1 CCGAAGAAACUCGAGCGGAGCUAGCGUU-UUCGUUGCUCCGGGGGAGUUAUCCGUACUCGAAGUGUCGUUCAAUAUAUUUGCAUAUAUUGUUAUACUUUUCGGAUUUGACAAUCAUAGUU (((((((...(((((((((((.((((..-..))))))))))..(((....)))...))))).((((....((((((((....))))))))..)))))))))))................. ( -37.90) >DroSim_CAF1 19471 119 + 1 CCGAAGAAACUCGAGCGGAGCUAGCGUU-UUCGUUGCUCCGGGGGAGUUAUCCGUACUCGAAGUGUCGUUCAAUAUAUUUGCAUAUAUUGUUAUACUUUUCGGAUUUGACAAUCAUAGUU (((((((...(((((((((((.((((..-..))))))))))..(((....)))...))))).((((....((((((((....))))))))..)))))))))))................. ( -37.90) >DroEre_CAF1 15879 119 + 1 CCGAAGAAACUCGAACGGAGCUGACGUU-UUCGAUGCUCUCCGUGAGUUAUCCGUACUCAAAGUGUCGCUCAAUAUAUUUGCAUAUAUUGUUAUAUUUUCCGGAUUUGACAAUCAUAGUU .((((..((((((...(((((...((..-..))..)))))...)))))).((((.....(((((((....((((((((....))))))))..))))))).))))))))............ ( -25.30) >DroYak_CAF1 16928 120 + 1 CCGAAGAAACUCGAACGGAGCUGGCGUUUUUCGAUGCUCCGACGGAGUUAUCCGUACUCGAAGUGUCGCUCAAUAUAUUUGCAUAUAUUGUUAUAUUUUUCGGAUUUGACAAUCAUAGUU ((((((((((((((.((((((...((.....))..))))))(((((....)))))..))).)))......((((((((....)))))))).....))))))))................. ( -36.40) >consensus CCGAAGAAACUCGAGCGGAGCUAGCGUU_UUCGUUGCUCCGGGGGAGUUAUCCGUACUCGAAGUGUCGUUCAAUAUAUUUGCAUAUAUUGUUAUACUUUUCGGAUUUGACAAUCAUAGUU .((((..(((((...((((((.((((.....))))))))))...))))).((((.....(((((((....((((((((....))))))))..))))))).))))))))............ (-30.84 = -30.64 + -0.20)



| Location | 15,816,485 – 15,816,602 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15816485 117 - 22224390 AACUAUGAAUGUCAAAUCCGAAAAGUAUAACAAUAUAUGCAAAUAUAUUGAACGACACUUCGAGUACGGAUAACUC--CCGGAGCAACGAA-AACGCUAGCUCCGCUCGAGUUUCUUCGG .................(((((........((((((((....))))))))...((.(((.(((((..(((....))--).(((((..((..-..))...))))))))))))).))))))) ( -31.90) >DroSec_CAF1 16808 119 - 1 AACUAUGAUUGUCAAAUCCGAAAAGUAUAACAAUAUAUGCAAAUAUAUUGAACGACACUUCGAGUACGGAUAACUCCCCCGGAGCAACGAA-AACGCUAGCUCCGCUCGAGUUUCUUCGG .................(((((........((((((((....))))))))...((.(((.(((((..(((....)))...(((((..((..-..))...))))))))))))).))))))) ( -31.90) >DroSim_CAF1 19471 119 - 1 AACUAUGAUUGUCAAAUCCGAAAAGUAUAACAAUAUAUGCAAAUAUAUUGAACGACACUUCGAGUACGGAUAACUCCCCCGGAGCAACGAA-AACGCUAGCUCCGCUCGAGUUUCUUCGG .................(((((........((((((((....))))))))...((.(((.(((((..(((....)))...(((((..((..-..))...))))))))))))).))))))) ( -31.90) >DroEre_CAF1 15879 119 - 1 AACUAUGAUUGUCAAAUCCGGAAAAUAUAACAAUAUAUGCAAAUAUAUUGAGCGACACUUUGAGUACGGAUAACUCACGGAGAGCAUCGAA-AACGUCAGCUCCGUUCGAGUUUCUUCGG .(((.....((((...(....)........((((((((....))))))))...)))).....))).((((.(((((((((((.....((..-..))....))))))..)))))..)))). ( -25.90) >DroYak_CAF1 16928 120 - 1 AACUAUGAUUGUCAAAUCCGAAAAAUAUAACAAUAUAUGCAAAUAUAUUGAGCGACACUUCGAGUACGGAUAACUCCGUCGGAGCAUCGAAAAACGCCAGCUCCGUUCGAGUUUCUUCGG .................(((((........((((((((....))))))))...((.(((.((((.(((((....)))))((((((..((.....))...))))))))))))).))))))) ( -32.10) >consensus AACUAUGAUUGUCAAAUCCGAAAAGUAUAACAAUAUAUGCAAAUAUAUUGAACGACACUUCGAGUACGGAUAACUCCCCCGGAGCAACGAA_AACGCUAGCUCCGCUCGAGUUUCUUCGG .....(((.......(((((..........((((((((....))))))))..(((....)))....)))))(((((...((((((..((.....))...))))))...)))))...))). (-26.96 = -27.20 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:29 2006