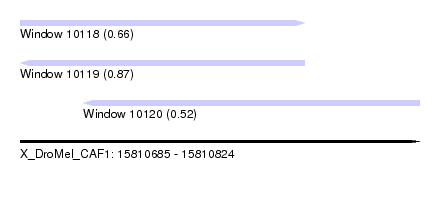
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,810,685 – 15,810,824 |
| Length | 139 |
| Max. P | 0.867841 |

| Location | 15,810,685 – 15,810,784 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -22.05 |
| Energy contribution | -21.39 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15810685 99 + 22224390 UGAUUUUCAAAAUUCGAAACUUUGACAAAUUUUGAAGCUGGGAUUGGAGCUCCUAGAGGUUAUCCCUCUCCUUGCACAGCCAGCGCUCCUAGUUGGCCA .....((((((((((((....))))...))))))))((.((((..(((...((....))...)))...)))).))...((((((.......)))))).. ( -27.70) >DroSec_CAF1 12406 90 + 1 UGAUUUCCAAGAUUUCAAACUUUGACCAAUUUUGGAGCUGGGAUUGGAGCUCCUAGAGAC---------ACUAGUACAGCCAGCUGGACUAGUUGGCCA ....(((((((((((((.....)))..))))))))))((((((.......))))))...(---------((((((.(((....))).))))).)).... ( -25.30) >DroSim_CAF1 12275 90 + 1 UGAUUUCCAAGAUUUCAAACUUUGACCAAUUUUGGAGCUGGGAUUGGAGCUCCUAGAGAC---------ACUAGUACAGCCAGCUGGACUAGUUGGCCA ....(((((((((((((.....)))..))))))))))((((((.......))))))...(---------((((((.(((....))).))))).)).... ( -25.30) >consensus UGAUUUCCAAGAUUUCAAACUUUGACCAAUUUUGGAGCUGGGAUUGGAGCUCCUAGAGAC_________ACUAGUACAGCCAGCUGGACUAGUUGGCCA ....((((((((((.............))))))))))((((((.......))))))......................((((((.......)))))).. (-22.05 = -21.39 + -0.66)

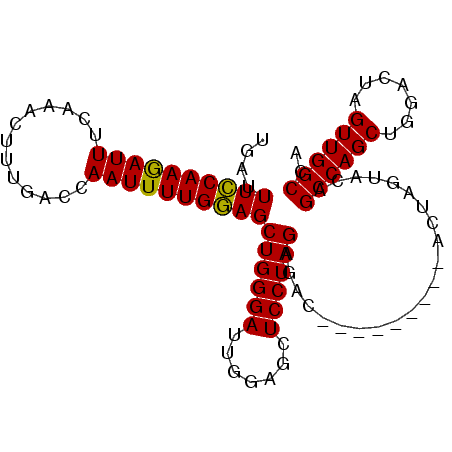
| Location | 15,810,685 – 15,810,784 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -19.35 |
| Energy contribution | -20.80 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15810685 99 - 22224390 UGGCCAACUAGGAGCGCUGGCUGUGCAAGGAGAGGGAUAACCUCUAGGAGCUCCAAUCCCAGCUUCAAAAUUUGUCAAAGUUUCGAAUUUUGAAAAUCA ..........(((((((.......))....(((((.....)))))....))))).........(((((((((((.........)))))))))))..... ( -27.50) >DroSec_CAF1 12406 90 - 1 UGGCCAACUAGUCCAGCUGGCUGUACUAGU---------GUCUCUAGGAGCUCCAAUCCCAGCUCCAAAAUUGGUCAAAGUUUGAAAUCUUGGAAAUCA ((((((((((((.(((....))).))))))---------.......((((((........)))))).....))))))...................... ( -27.70) >DroSim_CAF1 12275 90 - 1 UGGCCAACUAGUCCAGCUGGCUGUACUAGU---------GUCUCUAGGAGCUCCAAUCCCAGCUCCAAAAUUGGUCAAAGUUUGAAAUCUUGGAAAUCA ((((((((((((.(((....))).))))))---------.......((((((........)))))).....))))))...................... ( -27.70) >consensus UGGCCAACUAGUCCAGCUGGCUGUACUAGU_________GUCUCUAGGAGCUCCAAUCCCAGCUCCAAAAUUGGUCAAAGUUUGAAAUCUUGGAAAUCA ((((((((((((.(((....))).))))).................((((((........))))))....)))))))...................... (-19.35 = -20.80 + 1.45)



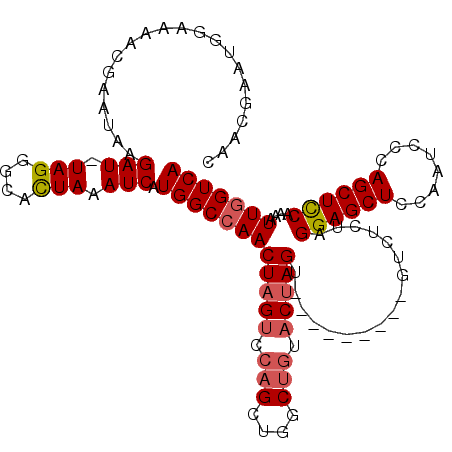
| Location | 15,810,707 – 15,810,824 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.16 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -21.97 |
| Energy contribution | -23.20 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15810707 117 - 22224390 UACCGAAUGAAAAACGAACCAGAUUUAGGGCAUUAAAUCAUGGCCAACUAGGAGCGCUGGCUGUGCAAGGAGAGGGAUAACCUCUAGGAGCUCCAAUCCCAGCUUCAAAAUUUGUCA ..((..............((((((((((....))))))).)))((.....)).((((.....))))..))(((((.....))))).((((((........))))))........... ( -29.10) >DroSec_CAF1 12428 107 - 1 CAACGAAUGGAAAACGAAUAAGAU-UAGGGCACUAAAUCAUGGCCAACUAGUCCAGCUGGCUGUACUAGU---------GUCUCUAGGAGCUCCAAUCCCAGCUCCAAAAUUGGUCA .....................(((-(((....)).)))).((((((((((((.(((....))).))))))---------.......((((((........)))))).....)))))) ( -29.90) >DroSim_CAF1 12297 107 - 1 CAACGAAUGGAAAACGAAUAAGAU-UAGGGCACUAAAUCAUGGCCAACUAGUCCAGCUGGCUGUACUAGU---------GUCUCUAGGAGCUCCAAUCCCAGCUCCAAAAUUGGUCA .....................(((-(((....)).)))).((((((((((((.(((....))).))))))---------.......((((((........)))))).....)))))) ( -29.90) >consensus CAACGAAUGGAAAACGAAUAAGAU_UAGGGCACUAAAUCAUGGCCAACUAGUCCAGCUGGCUGUACUAGU_________GUCUCUAGGAGCUCCAAUCCCAGCUCCAAAAUUGGUCA .....................(((.(((....))).))).((((((((((((.(((....))).))))).................((((((........))))))....))))))) (-21.97 = -23.20 + 1.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:22 2006