
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,808,366 – 15,808,555 |
| Length | 189 |
| Max. P | 0.807275 |

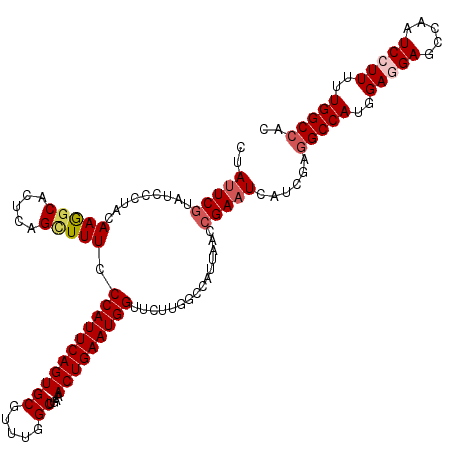
| Location | 15,808,366 – 15,808,486 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -28.98 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15808366 120 + 22224390 CUAUUCGUAUCCCUACAAAGCACUUAGCUUUCCCAUUCAGUGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCAUUAACCGAAUCAUCGAGGCCAUGGAGGAGCCAAUCCUUAUUGGCCAC ..(((((.........(((((.....))))).(((((((((((.....))....)))))))))...............)))))......(((((..(((((.....)))))..))))).. ( -34.60) >DroSec_CAF1 10274 120 + 1 CUAUUCGUAUCCCUACAAGGCACUCAGCUUUCCCAUUCAGUGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCAUUAACCGAAUCAUCGAGGCCAUGGAGGAGCCAAUCCUUUUUGGCCAC ..(((((........((((((.....))....(((((((((((.....))....)))))))))..)))).........)))))......(((((..(((((.....)))))..))))).. ( -36.83) >DroSim_CAF1 10156 120 + 1 CUAUUCGUAUCCCUACAAGGCACUCAGCUUUCCCAUUCAGUGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCAUUAACCGAAUCAUCGAUGCCAUGGAGGAGCCAAUCCUUUUUGGCCAC ...........(((.((.((((....(((...(((((((((((.....))....))))))))).....))).......(((....))).)))).)).))).(((((......)))))... ( -33.20) >DroEre_CAF1 8740 120 + 1 CUACUCGUAUCCCUACAAGGCGCUCAGUUUUCCCAUUCAGUGCGUUUGGCUGAAACUGAAUGGCUCCUGGCCAGUAAGCGAAUCAUCGAGGCCAUGGAAGAGGCAAUCCUUUUUGGCCAC ....(((....((.....))(((((((((((.(((..(.....)..)))..)))))))).((((.....))))....)))......)))(((((...((((((....))))))))))).. ( -34.40) >DroYak_CAF1 9594 120 + 1 CUAUUCAUAUCCCUACAAAGCACUCCGAUUUCCCAUUCAGUGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCACUAUCGGAAUCAUCGAGGCCAUGGAAGAGUGACUCCUUUUUGGCCAC .........(((.......(((((..((........)))))))...((((..(((((....)))).)..)))).....)))........(((((..((.(((...))).))..))))).. ( -31.30) >consensus CUAUUCGUAUCCCUACAAGGCACUCAGCUUUCCCAUUCAGUGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCAUUAACCGAAUCAUCGAGGCCAUGGAGGAGCCAAUCCUUUUUGGCCAC ..(((((.........(((((.....))))).(((((((((((.....))....)))))))))...............)))))......(((((..(((((.....)))))..))))).. (-28.98 = -29.82 + 0.84)


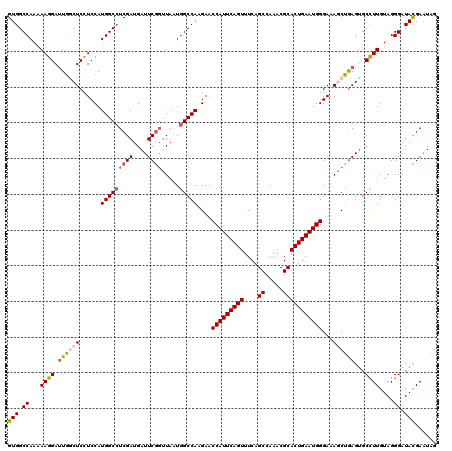
| Location | 15,808,366 – 15,808,486 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -30.32 |
| Energy contribution | -31.04 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15808366 120 - 22224390 GUGGCCAAUAAGGAUUGGCUCCUCCAUGGCCUCGAUGAUUCGGUUAAUGGCCAAGAACCAUUCAGUUUCAGCCAAACGCACUGAAUGGGAAAGCUAAGUGCUUUGUAGGGAUACGAAUAG (((.((.((((((.((((((..(((.(((((((((....)))).....))))).....((((((((....((.....))))))))))))).))))))...)))))).))..)))...... ( -38.50) >DroSec_CAF1 10274 120 - 1 GUGGCCAAAAAGGAUUGGCUCCUCCAUGGCCUCGAUGAUUCGGUUAAUGGCCAAGAACCAUUCAGUUUCAGCCAAACGCACUGAAUGGGAAAGCUGAGUGCCUUGUAGGGAUACGAAUAG (((.((...((((.(..(((..(((.(((((((((....)))).....))))).....((((((((....((.....))))))))))))).)))..)...))))....)).)))...... ( -38.70) >DroSim_CAF1 10156 120 - 1 GUGGCCAAAAAGGAUUGGCUCCUCCAUGGCAUCGAUGAUUCGGUUAAUGGCCAAGAACCAUUCAGUUUCAGCCAAACGCACUGAAUGGGAAAGCUGAGUGCCUUGUAGGGAUACGAAUAG (((.((...((((.(..(((..(((.(((((((((....))))).....)))).....((((((((....((.....))))))))))))).)))..)...))))....)).)))...... ( -36.80) >DroEre_CAF1 8740 120 - 1 GUGGCCAAAAAGGAUUGCCUCUUCCAUGGCCUCGAUGAUUCGCUUACUGGCCAGGAGCCAUUCAGUUUCAGCCAAACGCACUGAAUGGGAAAACUGAGCGCCUUGUAGGGAUACGAGUAG (((.((...((((...((.((((((.(((((.(((....)))......)))))))))(((((((((....((.....))))))))))).......)))).))))....)).)))...... ( -36.70) >DroYak_CAF1 9594 120 - 1 GUGGCCAAAAAGGAGUCACUCUUCCAUGGCCUCGAUGAUUCCGAUAGUGGCCAAGAACCAUUCAGUUUCAGCCAAACGCACUGAAUGGGAAAUCGGAGUGCUUUGUAGGGAUAUGAAUAG (((.((.....((((.((((((....((((((((.......)))....)))))....(((((((((....((.....)))))))))))......))))))))))....)).)))...... ( -34.70) >consensus GUGGCCAAAAAGGAUUGGCUCCUCCAUGGCCUCGAUGAUUCGGUUAAUGGCCAAGAACCAUUCAGUUUCAGCCAAACGCACUGAAUGGGAAAGCUGAGUGCCUUGUAGGGAUACGAAUAG (((.((...((((.((((((......(((((((((....)))).....)))))....(((((((((....((.....)))))))))))...))))))...))))....)).)))...... (-30.32 = -31.04 + 0.72)


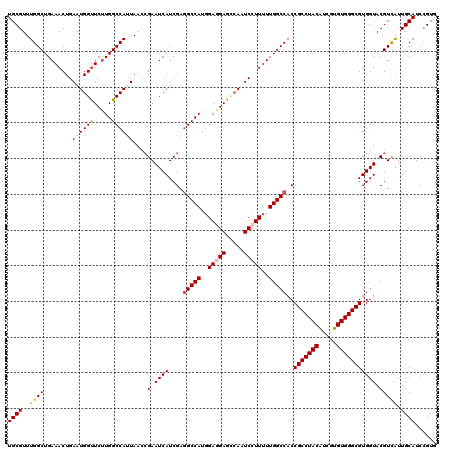
| Location | 15,808,406 – 15,808,526 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -42.28 |
| Consensus MFE | -38.40 |
| Energy contribution | -38.92 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15808406 120 + 22224390 UGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCAUUAACCGAAUCAUCGAGGCCAUGGAGGAGCCAAUCCUUAUUGGCCACCGCCUACAUCGUGUGGGCGUGGUACGUCAUUGCAUCUGUG ((((..((((........((((((.....)))))).((((.........(((((..(((((.....)))))..)))))..(((((((.....)))))))))))..)))).))))...... ( -43.80) >DroSec_CAF1 10314 120 + 1 UGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCAUUAACCGAAUCAUCGAGGCCAUGGAGGAGCCAAUCCUUUUUGGCCACCGCCUACAUCGUGUGGGCGUGGUACGUCAUUGCAUCCGUG ((((..((((........((((((.....)))))).((((.........(((((..(((((.....)))))..)))))..(((((((.....)))))))))))..)))).))))...... ( -45.50) >DroSim_CAF1 10196 120 + 1 UGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCAUUAACCGAAUCAUCGAUGCCAUGGAGGAGCCAAUCCUUUUUGGCCACCGCCUACAACGUGUGGGCGUGGUACGUCAUUGCAUCCGUG .(((..((((..(((((....)))).)..))))..............(((((((..(((((.....)))))..))(((((.((((((.....)))))))))))........)))))))). ( -43.00) >DroEre_CAF1 8780 120 + 1 UGCGUUUGGCUGAAACUGAAUGGCUCCUGGCCAGUAAGCGAAUCAUCGAGGCCAUGGAAGAGGCAAUCCUUUUUGGCCACCGCCUACAUAGUGUGGGCGUGGUACGUUGUUGCAUCCGUG ((((..((((..(((..((.((.(((((((((......(((....))).))))).)))...).)).))..)))..))))((((((((.....))))))).).........))))...... ( -40.30) >DroYak_CAF1 9634 120 + 1 UGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCACUAUCGGAAUCAUCGAGGCCAUGGAAGAGUGACUCCUUUUUGGCCACCGCCUACGUAAUGUGGGCGUGGUACGUCAUUGCAUCCGUU ......((((..(((((....)))).)..))))....((((..((..(((((((..((.(((...))).))..))))).((((((((.....))))))).).....))..))..)))).. ( -38.80) >consensus UGCGUUUGGCUGAAACUGAAUGGUUCUUGGCCAUUAACCGAAUCAUCGAGGCCAUGGAGGAGCCAAUCCUUUUUGGCCACCGCCUACAUCGUGUGGGCGUGGUACGUCAUUGCAUCCGUG ((((..((((........((((((.....))))))....(.((((....(((((..(((((.....)))))..)))))..(((((((.....))))))))))).))))).))))...... (-38.40 = -38.92 + 0.52)


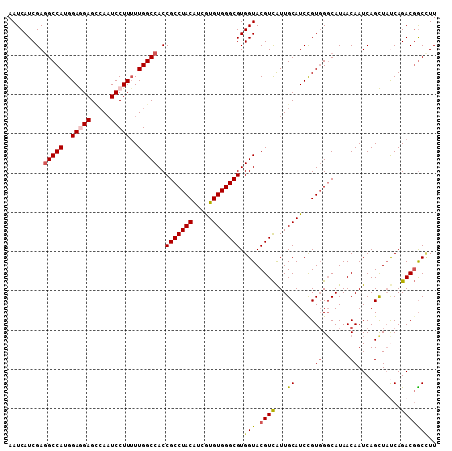
| Location | 15,808,446 – 15,808,555 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -29.68 |
| Energy contribution | -30.20 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15808446 109 + 22224390 AAUCAUCGAGGCCAUGGAGGAGCCAAUCCUUAUUGGCCACCGCCUACAUCGUGUGGGCGUGGUACGUCAUUGCAUCUGUGGGCAUAACAAUCAGCUAUCAGACGGCCUU .......((((((((((((((.....)))))....(((((.((((((.....))))))))))).))).......((((..(((..........)))..)))).)))))) ( -38.70) >DroSec_CAF1 10354 109 + 1 AAUCAUCGAGGCCAUGGAGGAGCCAAUCCUUUUUGGCCACCGCCUACAUCGUGUGGGCGUGGUACGUCAUUGCAUCCGUGGCCAUAACAAUCAGCUAUCAGACGGCCUU .......((((((..((((((.....)))))).(((((((.((((((.....))))))(..((.....))..)....)))))))...................)))))) ( -40.90) >DroSim_CAF1 10236 109 + 1 AAUCAUCGAUGCCAUGGAGGAGCCAAUCCUUUUUGGCCACCGCCUACAACGUGUGGGCGUGGUACGUCAUUGCAUCCGUGGGCAUAACAAUCAGCUAUCAGACGGCCUU ...(((.(((((((..(((((.....)))))..))(((((.((((((.....)))))))))))........))))).)))(((.....................))).. ( -36.10) >DroEre_CAF1 8820 109 + 1 AAUCAUCGAGGCCAUGGAAGAGGCAAUCCUUUUUGGCCACCGCCUACAUAGUGUGGGCGUGGUACGUUGUUGCAUCCGUGGGCAUAACAAUCAGUUUCCAGACCACUUU .........(.(((((((....(((((........(((((.((((((.....))))))))))).....))))).))))))).).......................... ( -35.12) >DroYak_CAF1 9674 109 + 1 AAUCAUCGAGGCCAUGGAAGAGUGACUCCUUUUUGGCCACCGCCUACGUAAUGUGGGCGUGGUACGUCAUUGCAUCCGUUGGCAUAACAAUCAGUUACCAGACGGCAUU ..........((((((((.(((((((..(.....)(((((.((((((.....)))))))))))..)))))).).)))))(((..((((.....)))))))...)))... ( -35.00) >consensus AAUCAUCGAGGCCAUGGAGGAGCCAAUCCUUUUUGGCCACCGCCUACAUCGUGUGGGCGUGGUACGUCAUUGCAUCCGUGGGCAUAACAAUCAGCUAUCAGACGGCCUU .........(((((..(((((.....)))))..)))))..(((((((.....))))))).(((.((((...((.....((.......))....)).....))))))).. (-29.68 = -30.20 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:16 2006