| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,798,655 – 15,798,751 |
| Length | 96 |
| Max. P | 0.768145 |

| Location | 15,798,655 – 15,798,751 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -17.63 |
| Energy contribution | -19.13 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
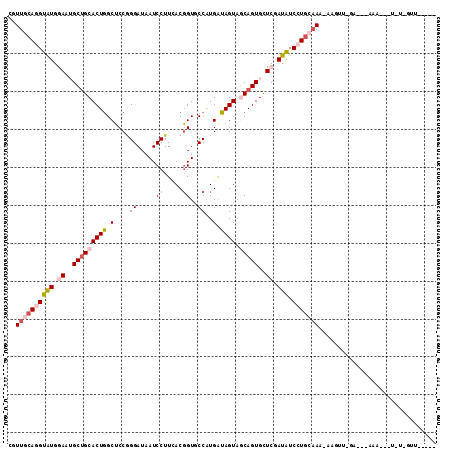
>X_DroMel_CAF1 15798655 96 + 22224390 CGUUGCAGGUAUGGAAUGCUGCACUGGCUCCGGGAUAAUCCUUUACGGUGCCAUGAUAGUAGCAGUGCUCGAUAUCCUGCAAA-GAGUU--------------UAGUUU--UU ..((((((((((.((..(((((((((((.(((((.....))....))).))).....))).)))))..)).))).))))))).-.....--------------......--.. ( -34.60) >DroGri_CAF1 3279 98 + 1 CGUUGCAAGUAUGAAAGGCAGCACUGGCACCAGGAUAAUCCUUGACUGUACCAUGAUAGUAACAGUGUUCGAUGUCCUGGAAA-AAAAAAGA---CAAA--U-C-A------- .(((((...........))))).......((((((((.((....(((((((.......)).)))))....))))))))))...-........---....--.-.-.------- ( -22.70) >DroSec_CAF1 2470 109 + 1 CGUUGCAGGUAUGGAAUGCUGCACUGGCUCCGGGAUAAUCCUUGACGGUGCCGUGAUAGUAGCAGUGCUCGAUAUCCUGCAAA-GAGUU-GA-GCAAGCCAUUU-GUUAAUUU ..((((((((((.((..(((((((((((.(((((.....))....))).))).....))).)))))..)).))).))))))).-(((((-((-.((((...)))-)))))))) ( -36.70) >DroEre_CAF1 2439 107 + 1 CGUUGCAGGUGUGGAAUGCUGCACUGGCUCCGGGAUAAUCCUUCACGGUCCCGUGGUAGUAGCAGUGCUCGAUAUCCUGCAAA-AAGUC-GAU-CAAAGAGGUUAGUUCA--- ..((((((((((.((..(((((((((.(..((((((...........))))))..))))).)))))..)).))).))))))).-.....-(((-(.....))))......--- ( -38.90) >DroWil_CAF1 2264 94 + 1 CGUUGCAGGUAUGAAAAGCUGCACUAGCUCCUGGAUAAUCCUUUACGGUUCCAUGAUAGUAACAGUGCUCAAUAUCCUAAUAAAAAUAU-GC---AAA--------------- ..((((((((((.....((((.((((..((.((((....((.....)).)))).))))))..)))).....)))))............)-))---)).--------------- ( -18.00) >DroYak_CAF1 2440 107 + 1 CGUUGCACGUGUGGAAUGCUGCACUGGCUCCGGGAUAAUCCUUCACGGUGCCGUGGUAGUAGCAGUGCUCGAUAUCCUGCAAC-AAGUC-AAU-CCAAUGGUUUAGUUCA--- .((((((.((((.((..(((((((((((.((((((......))).))).))).....))).)))))..)).))))..))))))-((..(-(..-....))..))......--- ( -33.40) >consensus CGUUGCAGGUAUGGAAUGCUGCACUGGCUCCGGGAUAAUCCUUCACGGUGCCAUGAUAGUAGCAGUGCUCGAUAUCCUGCAAA_AAGUU_GA___AAA___U_U_GUU_____ ..((((((((((.((..(((((((((.(....((.....((.....))..))..).)))).)))))..)).))).)))))))............................... (-17.63 = -19.13 + 1.50)


| Location | 15,798,655 – 15,798,751 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -18.05 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
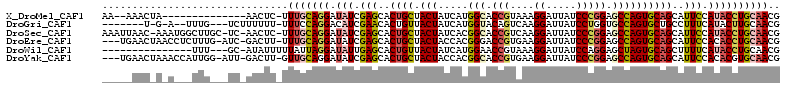
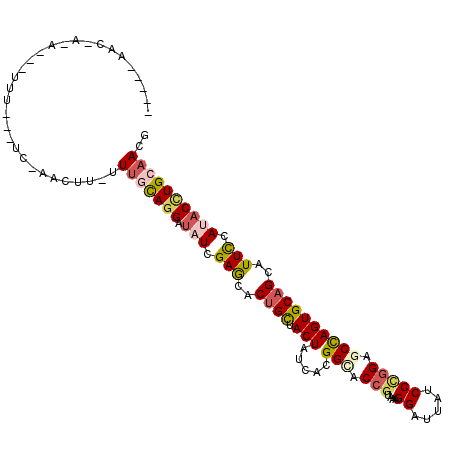
>X_DroMel_CAF1 15798655 96 - 22224390 AA--AAACUA--------------AACUC-UUUGCAGGAUAUCGAGCACUGCUACUAUCAUGGCACCGUAAAGGAUUAUCCCGGAGCCAGUGCAGCAUUCCAUACCUGCAACG ..--......--------------.....-.(((((((.(((.(((..((((.(((.....(((.(((....((.....))))).))))))))))..))).)))))))))).. ( -29.10) >DroGri_CAF1 3279 98 - 1 -------U-G-A--UUUG---UCUUUUUU-UUUCCAGGACAUCGAACACUGUUACUAUCAUGGUACAGUCAAGGAUUAUCCUGGUGCCAGUGCUGCCUUUCAUACUUGCAACG -------.-.-.--....---........-...((((((.(((....(((((.(((.....))))))))....)))..))))))(((.((((.((.....)))))).)))... ( -20.50) >DroSec_CAF1 2470 109 - 1 AAAUUAAC-AAAUGGCUUGC-UC-AACUC-UUUGCAGGAUAUCGAGCACUGCUACUAUCACGGCACCGUCAAGGAUUAUCCCGGAGCCAGUGCAGCAUUCCAUACCUGCAACG ........-...........-..-.....-.(((((((.(((.(((..((((.(((.....(((.(((....((.....))))).))))))))))..))).)))))))))).. ( -29.10) >DroEre_CAF1 2439 107 - 1 ---UGAACUAACCUCUUUG-AUC-GACUU-UUUGCAGGAUAUCGAGCACUGCUACUACCACGGGACCGUGAAGGAUUAUCCCGGAGCCAGUGCAGCAUUCCACACCUGCAACG ---................-...-.....-.(((((((.....(((..((((.(((.(..(((((((.....))....)))))..)..)))))))..)))....))))))).. ( -28.20) >DroWil_CAF1 2264 94 - 1 ---------------UUU---GC-AUAUUUUUAUUAGGAUAUUGAGCACUGUUACUAUCAUGGAACCGUAAAGGAUUAUCCAGGAGCUAGUGCAGCUUUUCAUACCUGCAACG ---------------.((---((-(((((((....))))))....((((((......((.((((.((.....))....)))).))..)))))).............))))).. ( -17.80) >DroYak_CAF1 2440 107 - 1 ---UGAACUAAACCAUUGG-AUU-GACUU-GUUGCAGGAUAUCGAGCACUGCUACUACCACGGCACCGUGAAGGAUUAUCCCGGAGCCAGUGCAGCAUUCCACACGUGCAACG ---....((((....))))-...-.....-(((((((((...(..((((((((.((..((((....))))..((......)))))).)))))).)...))).....)))))). ( -27.80) >consensus _____AAC_A_A___UUU___UC_AACUU_UUUGCAGGAUAUCGAGCACUGCUACUAUCACGGCACCGUAAAGGAUUAUCCCGGAGCCAGUGCAGCAUUCCAUACCUGCAACG ...............................(((((((.(((.(((..((((.(((.....(((.(((....((.....))))).))))))))))..))).)))))))))).. (-18.05 = -18.88 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:08 2006