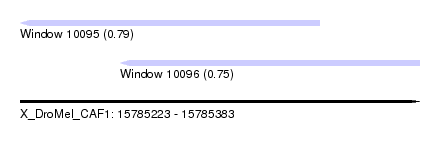
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,785,223 – 15,785,383 |
| Length | 160 |
| Max. P | 0.789714 |

| Location | 15,785,223 – 15,785,343 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -49.60 |
| Consensus MFE | -46.19 |
| Energy contribution | -45.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
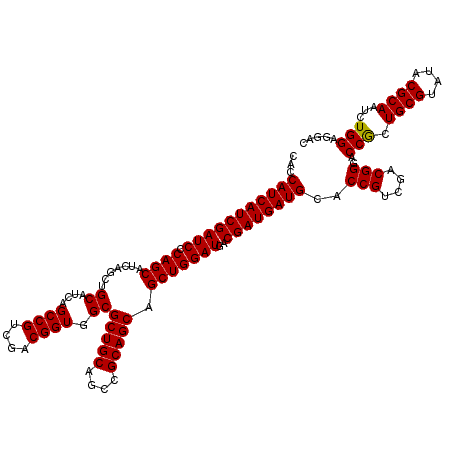
>X_DroMel_CAF1 15785223 120 - 22224390 CACCAUCAUCGAUCCCAGCAUCAAUUGCAUCAGCCGUCGACGGUGGCGCUGCAGCCGCAGCAGCUGGAUGACGAUGAUGCACCGUCGACGGGACCACUGCGUAUACGCAAUCUGGAGGAC ...((((((((.(((((((.......((....((((....)))).))(((((....))))).)))))..))))))))))..(((....)))..(((.((((....))))...)))..... ( -48.10) >DroSim_CAF1 16957 120 - 1 CACCAUCAUCGAUCCCAGCACCAGCUGCAUCAGCCGUCGACGGUGGCGCUGCAGCCGCAGCAGCUGGAUGACGAUGAUGCACCGUCGACGGGACCGCUGCGUAUACGCAAUCUGGAGGAC ..((..((.((.((((..((((((((((....((((....))))(((......)))...)))))))).)).(((((......)))))..)))).)).((((....))))...))..)).. ( -49.00) >DroYak_CAF1 14660 120 - 1 CAUCAUCAUCGAUCGCAGCAUCAGCUGCAUCAGCCGUCGACGGUGGCGCUGCAGCCGCAGCAGCUGGAUGACGAUGAUGCACCGUCGACGGGACCGCUGCGUAUACGCAAUCUGGAGGAU ....(((.(((((.(((((....))))))))..(((((((((((((((((((....))))).))((.....))......))))))))))))))(((.((((....))))...)))..))) ( -51.70) >consensus CACCAUCAUCGAUCCCAGCAUCAGCUGCAUCAGCCGUCGACGGUGGCGCUGCAGCCGCAGCAGCUGGAUGACGAUGAUGCACCGUCGACGGGACCGCUGCGUAUACGCAAUCUGGAGGAC ...(((((((((((.((((.......((....((((....)))).))(((((....))))).)))))))..))))))))..(((....)))..(((.((((....))))...)))..... (-46.19 = -45.97 + -0.22)


| Location | 15,785,263 – 15,785,383 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -54.57 |
| Consensus MFE | -50.30 |
| Energy contribution | -50.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
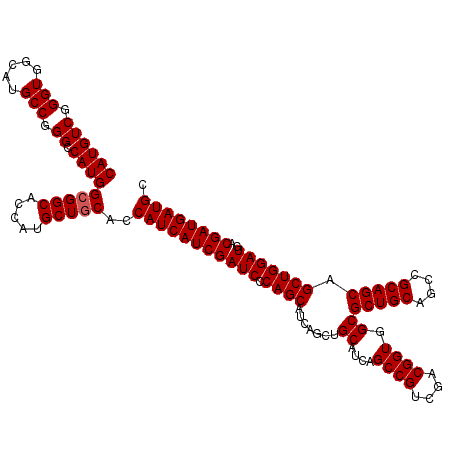
>X_DroMel_CAF1 15785263 120 - 22224390 CAUGUCGGGUGGCAUGCCGGGCCAUGGCGGCACCAUGCUGCACCAUCAUCGAUCCCAGCAUCAAUUGCAUCAGCCGUCGACGGUGGCGCUGCAGCCGCAGCAGCUGGAUGACGAUGAUGC ((((((.(((.....))).)).))))(((((.....)))))..((((((((.(((((((.......((....((((....)))).))(((((....))))).)))))..)))))))))). ( -54.50) >DroSim_CAF1 16997 120 - 1 CAUGUCGGGUGGGAUGCCGGGCCAUGGCGGCACCAUGCUGCACCAUCAUCGAUCCCAGCACCAGCUGCAUCAGCCGUCGACGGUGGCGCUGCAGCCGCAGCAGCUGGAUGACGAUGAUGC ((((((.(((.....))).)).))))(((((.....)))))..((((((((.(((((((....(((((....((((....))))(((......)))))))).)))))..)))))))))). ( -55.80) >DroYak_CAF1 14700 120 - 1 CAUGUCGGGUGGCAUGCCUGGCCAUGGCGGCACCAUGCUCCAUCAUCAUCGAUCGCAGCAUCAGCUGCAUCAGCCGUCGACGGUGGCGCUGCAGCCGCAGCAGCUGGAUGACGAUGAUGC ...(((((((.....))))))).((((.(((.....)))))))((((((((.(((((((....)))))(((.((((....))))((((((((....))))).))).))))))))))))). ( -53.40) >consensus CAUGUCGGGUGGCAUGCCGGGCCAUGGCGGCACCAUGCUGCACCAUCAUCGAUCCCAGCAUCAGCUGCAUCAGCCGUCGACGGUGGCGCUGCAGCCGCAGCAGCUGGAUGACGAUGAUGC ((((((.(((.....))).)).))))(((((.....)))))..(((((((((((.((((.......((....((((....)))).))(((((....))))).)))))))..)))))))). (-50.30 = -50.63 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:59 2006