
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,777,740 – 15,777,919 |
| Length | 179 |
| Max. P | 0.996024 |

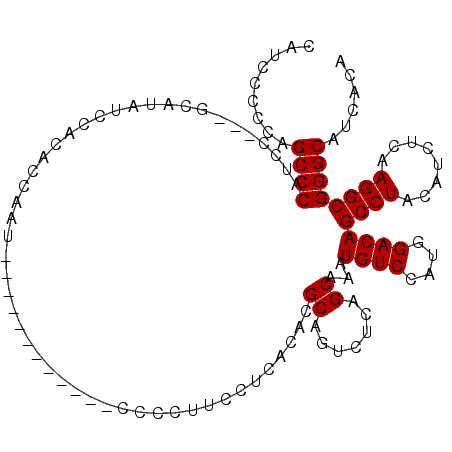
| Location | 15,777,740 – 15,777,839 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15777740 99 + 22224390 CAUCCCCCAGCCCAUCC---GCAUAUCCACACCAAU------------CCCCUUCCUCACACGCAGUCUCAGCAAAUGUCCAUGGACAGCCUACAUCUCAAGGCGGGCAUCGCA .........((((....---................------------..............((.......))...((((....))))((((........))))))))...... ( -20.10) >DroSec_CAF1 8323 99 + 1 CAUCCCCCAGCCCAUCC---GCAUAUCCACACCAAU------------CCCCUUCCUCACACGCAGUCUCAGCAAAUGUCCAUGGACAGCCUACAUCUCAAGGCGGGCAUCACA .........((((....---................------------..............((.......))...((((....))))((((........))))))))...... ( -20.10) >DroSim_CAF1 6763 99 + 1 CAUCCCCCAGCCCAUCC---GCAUAUCCACACCAAU------------CCCCUUCCUCACACGCAGUCUCAGCAAAUGUCCAUGGACAGCCUACAUCUCAAGGCGGGCAUCACA .........((((....---................------------..............((.......))...((((....))))((((........))))))))...... ( -20.10) >DroYak_CAF1 7994 114 + 1 CAUCCCCAAGCCCAUCCGCCGCAUAUCCAUCCCAAUCCCAAUGCCCUUCCUCUUCCACACACGCAGUCUCAGCAAAUGUCCUUGGACAGCCUACAUCUAAAGGCGGGCAUCACA .........((((.......((((................))))..................((.......))...((((....))))((((........))))))))...... ( -20.89) >consensus CAUCCCCCAGCCCAUCC___GCAUAUCCACACCAAU____________CCCCUUCCUCACACGCAGUCUCAGCAAAUGUCCAUGGACAGCCUACAUCUCAAGGCGGGCAUCACA .........((((.................................................((.......))...((((....))))((((........))))))))...... (-20.10 = -20.10 + -0.00)

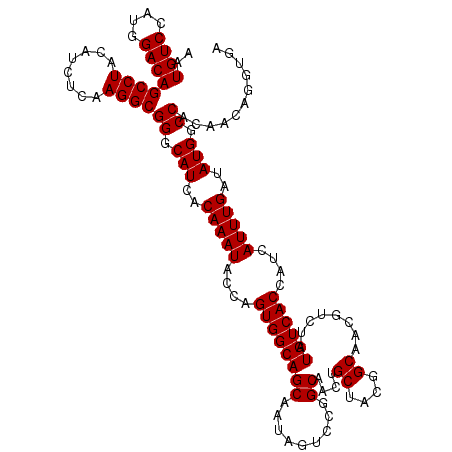
| Location | 15,777,799 – 15,777,919 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -29.94 |
| Energy contribution | -30.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15777799 120 + 22224390 AAUGUCCAUGGACAGCCUACAUCUCAAGGCGGGCAUCGCAAAUACCAGUGGCAGCAAUAGUCCGGCAACUGCAACGGCAACGUCUAUGUCACCAUCAUUUGAUAUGGCCACAACAGGUGA ..((((....))))((((........))))((.(((..(((((....(((((((((.......(....)))).(((....)))...))))))....)))))..))).))........... ( -33.21) >DroSec_CAF1 8382 120 + 1 AAUGUCCAUGGACAGCCUACAUCUCAAGGCGGGCAUCACAAAUACCAGUGGCAGCAAUAGUCCGGCAACAGCUACGGCAACGUCUAUGUCACCAUCAUUUGAUAUGGCCACAACAGGCGA ..((((....))))((((............((............)).(((((.((..((((..(....).))))..))......(((((((........))))))))))))...)))).. ( -33.40) >DroSim_CAF1 6822 120 + 1 AAUGUCCAUGGACAGCCUACAUCUCAAGGCGGGCAUCACAAAUACCAGUGGCAGCAAUAGUCCGGCAACAGCUACGGCAACGUCUAUGUCACCAUCAUUUGAUAUGGCCACAACAGGUGA ..((((....))))(((..(........)..)))........((((.(((((.((..((((..(....).))))..))......(((((((........))))))))))))....)))). ( -33.70) >DroEre_CAF1 8426 120 + 1 UAUGUCCAUGGACAGCCUACAUCUAAAGGCGGGCAUCACAAAUACCAGUGGCAGCAAUAGUCCCGCAACUGCUACAGCAACGUCUAUGUCACCAUCAUUUGAUAUGGCCACAACAGGUGA ..((((....))))(((..(........)..)))........((((.(((((((..............)))))))......(.((((((((........)))))))).)......)))). ( -31.34) >DroYak_CAF1 8068 120 + 1 AAUGUCCUUGGACAGCCUACAUCUAAAGGCGGGCAUCACAAAUACCAGUGGCAGCAAUAGUCCAGCAACUGCUACGGCAACGUCUAUGUCACCAUCAUUUGAUAUGGCCACAACAGGUGA ..((((....))))((((........))))((.(((..(((((....((((((..........(((....)))(((....)))...))))))....)))))..))).))........... ( -32.70) >consensus AAUGUCCAUGGACAGCCUACAUCUCAAGGCGGGCAUCACAAAUACCAGUGGCAGCAAUAGUCCGGCAACUGCUACGGCAACGUCUAUGUCACCAUCAUUUGAUAUGGCCACAACAGGUGA ..((((....))))((((........))))((.(((..(((((....((((((...........((....)).(((....)))...))))))....)))))..))).))........... (-29.94 = -30.14 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:53 2006