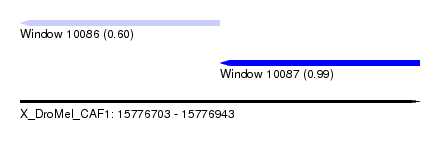
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,776,703 – 15,776,943 |
| Length | 240 |
| Max. P | 0.994748 |

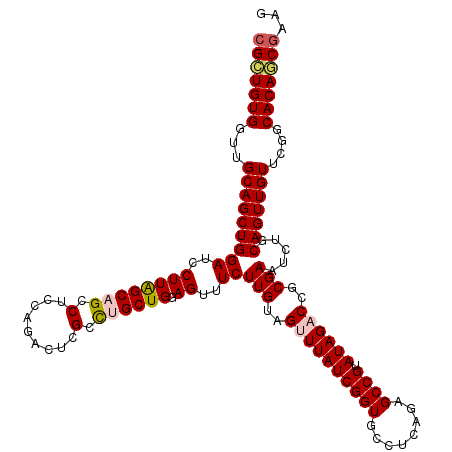
| Location | 15,776,703 – 15,776,823 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -46.48 |
| Consensus MFE | -37.32 |
| Energy contribution | -37.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15776703 120 - 22224390 CGCUGUGGUUGCAGCUGGAUCCUUAGCAGCCUCGAGACUCGCCUGCUGGAGUUUCUUGUAGUUUAUCGGUGCCUCAGAGCCGUAUAGACCGCGAAUCUGCAGUUGUUCGGCACAGCGAAG (((((((.(.(((((((((..((((((((...((.....)).))))).)))..))(((..((((((((((........)))).))))))..))).....)))))))..).)))))))... ( -45.80) >DroSec_CAF1 7297 120 - 1 CGCUGUGGUUGCAGCUGGAUCCUUAGCUGCCUCCAGACUAGCCUGCUGCAGUUUCUUGUAGUUUAUCGGUGCCUCAGAGCCGUAUAGACCGCGAAUCUGCAGUUGUUCGGCACAGCGAAG ((((((((..((((((((....))))))))..))......(((.(((((((....(((..((((((((((........)))).))))))..)))..))))))).....)))))))))... ( -52.50) >DroSim_CAF1 5734 120 - 1 CGCUGUGGUUGCAGCUGGAUCCUUAGCUGCCUCCAGACUAGCCUGCUGCAGUUUCUUGUAGUUUAUCGGUGCCUCAGAGCCGUAUAGACCGCGAAUCUGCAGUUGUUCGGCACAGCGAAG ((((((((..((((((((....))))))))..))......(((.(((((((....(((..((((((((((........)))).))))))..)))..))))))).....)))))))))... ( -52.50) >DroEre_CAF1 7316 120 - 1 GGGUGUGGAUGCAGCUGGAUCCUUGGCAGCCUCCAGACUCGCUUGCUGGAGUUUCUUGUAGUUUAUCGGUGCCUCGGAGCCGUAUAGCCCGCGAAUCUGCAGUUGUUCGGCACACCGAAG ((((.((((.((.((..(....)..)).)).)))).))))..((((.((.....)).))))....(((((((....).((((.(((((..(((....))).))))).)))).)))))).. ( -41.80) >DroYak_CAF1 6963 120 - 1 CGGUGUGGAUGCAGCUGGAUCCUUGGCAGCCUCCAGACUCGCCUGCUGGAGUUUCUUGUAGUUUAUCGGUGCCUCGGAGCCGUAUAGCCCGCGAAUCUGCAGUUGUUCGGCACAGCGAAG ..((.((((.((.((..(....)..)).)).)))).))((((.(((((((.....((((((.....(((.((..((....))....))))).....))))))...)))))))..)))).. ( -39.80) >consensus CGCUGUGGUUGCAGCUGGAUCCUUAGCAGCCUCCAGACUCGCCUGCUGGAGUUUCUUGUAGUUUAUCGGUGCCUCAGAGCCGUAUAGACCGCGAAUCUGCAGUUGUUCGGCACAGCGAAG (((((((...(((((((((..((((((((.(.........).)))))).))..))(((..((((((((((........)))).))))))..))).....)))))))....)))))))... (-37.32 = -37.88 + 0.56)


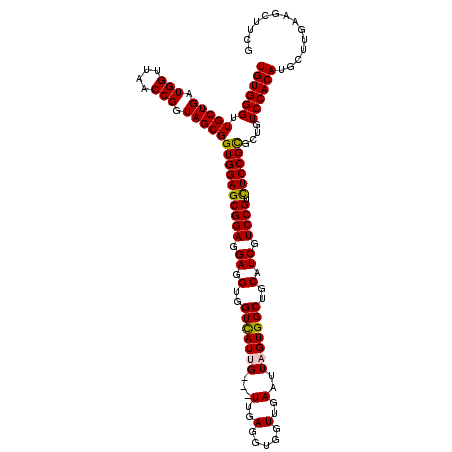
| Location | 15,776,823 – 15,776,943 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.91 |
| Mean single sequence MFE | -44.86 |
| Consensus MFE | -42.16 |
| Energy contribution | -41.92 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15776823 120 - 22224390 UGUGGGUUGCUGAUGGUUAACCGGUAGCGGUGGAGCGGAGGAGGUGGUCAUUGUUGUUGAGGUGGUUGAAUUAGUGGCUGCAUCGUCCGUUUCCGCGCUGUCCACAUGCUUGAAGCUUCG ((((((.(((((.(((....))).)))))((((((((((.((.(..(((((((...(..(.....)..)..)))))))..).)).)))).))))))....)))))).((.....)).... ( -45.50) >DroSec_CAF1 7417 117 - 1 UGUGGGUUGCUGAUGGUUAACCGGUAGCGGUGGAGCGGAGGAGGUGGUCAUUG---UUGAGGUGGUUGAAUUAGUGGCUGCAUCGUCCGUCUCCGCGCUGUCCACAUGCUUAAAACUUCG ((((((.(((((.(((....))).)))))((((((((((.((.(..(((((((---((.((....)).)).)))))))..).)).)))).))))))....)))))).............. ( -45.80) >DroSim_CAF1 5854 117 - 1 UGUGGGUUGCUGAUGGUUAACCGGUAGCGGUGGAGCGGAGGAGGUGGUCAUUG---UUGAGGUGGUUGAAUUAGUGGCUGCAUCGUCCGUCUCCGCGCUGUCCACAUGCUUGAAACUUCG ((((((.(((((.(((....))).)))))((((((((((.((.(..(((((((---((.((....)).)).)))))))..).)).)))).))))))....)))))).............. ( -45.80) >DroEre_CAF1 7436 117 - 1 UGUGGGGUGCUGCUGGUUAGCCGCUAGCGGUGGAGCGGAUGAGGUGGUUAUCG---UUGAGGUGGUAGAAUUAGUGGCUGCAUCGUCCGUCUCCGUGCUGUCCACAUGCUUGAAGCUUCG ((((((..(((((((((.....)))))))))(((((((((((.(..(((((..---.................)))))..).))))))).))))......)))))).((.....)).... ( -44.01) >DroYak_CAF1 7083 117 - 1 UGUGGGUUGCUGCUGGUUAACCGCUAGCGGUGGAGCGGAGGAGGUGGUUAUCG---UUGAGGUGGUUGAAUUAGUGGCUGCAUCGUCCGUCUCCGUGCUGUCCACAUGCUUGAAGCUUCG ((((((..(((((((((.....)))))))))((((((((.((.(..(((((..---(..(.....)..)....)))))..).)).)))).))))......)))))).((.....)).... ( -43.20) >consensus UGUGGGUUGCUGAUGGUUAACCGGUAGCGGUGGAGCGGAGGAGGUGGUCAUUG___UUGAGGUGGUUGAAUUAGUGGCUGCAUCGUCCGUCUCCGCGCUGUCCACAUGCUUGAAGCUUCG ((((((.(((((.(((....))).)))))((((((((((.((.(..(((((((...(..(.....)..)..)))))))..).)).)))).))))))....)))))).............. (-42.16 = -41.92 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:50 2006