
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,775,331 – 15,775,530 |
| Length | 199 |
| Max. P | 0.973373 |

| Location | 15,775,331 – 15,775,451 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -25.48 |
| Energy contribution | -25.96 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15775331 120 - 22224390 GGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUGAAUGUAUCAGCGCCGGAGUCUUGUUUAAAAAUACACACUAUUCUUGCAUCAAAUCGUGCACAACACAAAAAAAUAUGUACAUUUCA ((((.....))))((((((((((((((..((((....)))).)))((.(((((..(((.((....)).)))..))))).)).)))...))))))))........................ ( -28.90) >DroSec_CAF1 5890 120 - 1 GGACUGUAUGUCCUGUGCGCGUGAGCUAUGUACGAAUGUAUCAGCGCCAGAGUCUUGUUUAAAAAUACACACUAUUCUUGCAUUAAAUCGUGCACAACACAAAAAAAAAUGUACAAUUCA ((((.....))))(((((((....))..(((((((..((....))((.(((((..(((.((....)).)))..))))).))......)))))))................)))))..... ( -30.50) >DroSim_CAF1 4324 120 - 1 GGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUGAAUGUAUCAGCGCCAGAGUCUUGUUUAAAAAUACACACUAUUCUUGCAUUAAAUCGUGCACAACACAAAAAAAUAUGUACAAUUCA ((((.....))))(((((((....))..(((((((..((....))((.(((((..(((.((....)).)))..))))).))......)))))))................)))))..... ( -28.70) >DroEre_CAF1 5979 120 - 1 GGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUCAAUGUAUCAGCGCCAGAGUCUUCUUUAAAAAUACACACUAUUCUUGCAUCAAAUCGUUCACAGCACACACGAAUAUGUACAAUUCG ((((.....))))(((((..((((((...........((....))((.(((((....................))))).))........)))))).)))))..(((((.......))))) ( -26.05) >DroYak_CAF1 5589 120 - 1 GGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUGAAUGUAUCAGCGCCAGAGUCUUGUUUAAAAAUACACACUAUUCUUGCACCAAAUCGUUCACAACACACACAAAUAUGUACAUUUCG ((((.....))))(((((((....))..(((.((((((.....(.((.(((((..(((.((....)).)))..))))).)).).....))))))..)))...........)))))..... ( -28.10) >consensus GGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUGAAUGUAUCAGCGCCAGAGUCUUGUUUAAAAAUACACACUAUUCUUGCAUCAAAUCGUGCACAACACAAAAAAAUAUGUACAAUUCA ((((.....))))(((((((....))..(((((((..((....))((.(((((..(((.((....)).)))..))))).))......)))))))................)))))..... (-25.48 = -25.96 + 0.48)

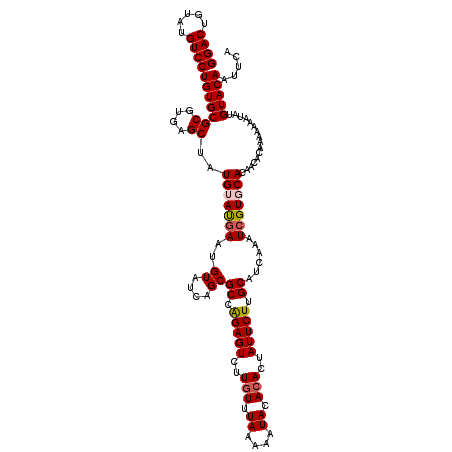
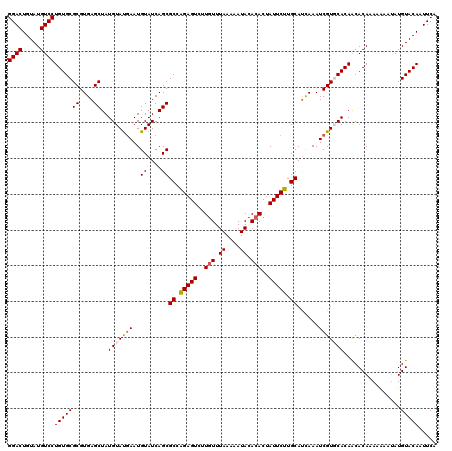
| Location | 15,775,411 – 15,775,530 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -22.78 |
| Energy contribution | -23.14 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15775411 119 + 22224390 UACAUUCAUACAUAGCUCACGCGCACAGGACAUACAGUCCUAACAUAA-AAUUCUCGUUCGUUUUCACGCCAUCGCAUCUAUUGUGUUACAUAGCGCCCAAAAAACGAGAAGUAAAGAAC ........(((...((......))..(((((.....))))).......-..((((((((..((....(((...((((.....)))).......)))...))..)))))))))))...... ( -22.50) >DroSec_CAF1 5970 119 + 1 UACAUUCGUACAUAGCUCACGCGCACAGGACAUACAGUCCUAACAUAA-AAUUCUCGUUCGUUUUCACGCCAUCGCAUCUAUUGUGUUACAUAGCGCCCAAAAAGCGAGAAAUAAAGUGC ..............((....))(((((((((.....))))).......-..((((((((..((....(((...((((.....)))).......)))...))..)))))))).....)))) ( -26.60) >DroSim_CAF1 4404 119 + 1 UACAUUCAUACAUAGCUCACGCGCACAGGACAUACAGUCCUAACAUAA-AAUUCUCGUUCGUUUUCACGCCAUCGCAUCUAUUGUGUUACAUAGCGCCCAAAAAGCGAGAAGUAAAGUGC ..............((....))(((((((((.....))))).......-..((((((((..((....(((...((((.....)))).......)))...))..)))))))).....)))) ( -26.60) >DroEre_CAF1 6059 120 + 1 UACAUUGAUACAUAGCUCACGCGCACAGGACAUACAGUCCUAACAUAAAAAUUCUCGUUCGUUUUCACGCCAUCGCAUCUAUUGUGUUACAUAGCGGCCAAAAAACGAGAAGUAAAGAGC ..............((((........(((((.....)))))..........((((((((..((....(((...((((.....)))).......)))...))..)))))))).....)))) ( -26.70) >DroYak_CAF1 5669 119 + 1 UACAUUCAUACAUAGCUCACGCGCACAGGACAUACAGUCCUAACAUAA-AAUUCUCGUUCGUUUUCACGCCAUCGCAUCUAUUGUGUUACAUAGCGGCCAAAAAGCGAGAAGUAAAGAGC ..............((((........(((((.....))))).......-..((((((((..((....(((...((((.....)))).......)))...))..)))))))).....)))) ( -26.70) >consensus UACAUUCAUACAUAGCUCACGCGCACAGGACAUACAGUCCUAACAUAA_AAUUCUCGUUCGUUUUCACGCCAUCGCAUCUAUUGUGUUACAUAGCGCCCAAAAAGCGAGAAGUAAAGAGC ..............((((........(((((.....)))))..........((((((((..((....(((...((((.....)))).......)))...))..)))))))).....)))) (-22.78 = -23.14 + 0.36)



| Location | 15,775,411 – 15,775,530 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -29.12 |
| Energy contribution | -28.96 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15775411 119 - 22224390 GUUCUUUACUUCUCGUUUUUUGGGCGCUAUGUAACACAAUAGAUGCGAUGGCGUGAAAACGAACGAGAAUU-UUAUGUUAGGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUGAAUGUA ((((..(((((((((((((((..((((((((((.(......).))).)))))))..))).)))))))))..-....((((((((.....)))))).))((....))...))).))))... ( -37.00) >DroSec_CAF1 5970 119 - 1 GCACUUUAUUUCUCGCUUUUUGGGCGCUAUGUAACACAAUAGAUGCGAUGGCGUGAAAACGAACGAGAAUU-UUAUGUUAGGACUGUAUGUCCUGUGCGCGUGAGCUAUGUACGAAUGUA ((((.....((((((.(((((..((((((((((.(......).))).)))))))..))).)).))))))..-.......(((((.....)))))))))((....)).............. ( -34.80) >DroSim_CAF1 4404 119 - 1 GCACUUUACUUCUCGCUUUUUGGGCGCUAUGUAACACAAUAGAUGCGAUGGCGUGAAAACGAACGAGAAUU-UUAUGUUAGGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUGAAUGUA ((((.....((((((.(((((..((((((((((.(......).))).)))))))..))).)).))))))..-.......(((((.....)))))))))((....)).............. ( -34.70) >DroEre_CAF1 6059 120 - 1 GCUCUUUACUUCUCGUUUUUUGGCCGCUAUGUAACACAAUAGAUGCGAUGGCGUGAAAACGAACGAGAAUUUUUAUGUUAGGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUCAAUGUA ((((.....(((((((((.((..((((((((((.(......).))).)))))).)..)).))))))))).....((((((((((.....))))))...)))))))).............. ( -35.20) >DroYak_CAF1 5669 119 - 1 GCUCUUUACUUCUCGCUUUUUGGCCGCUAUGUAACACAAUAGAUGCGAUGGCGUGAAAACGAACGAGAAUU-UUAUGUUAGGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUGAAUGUA ((((.....((((((.((.((..((((((((((.(......).))).)))))).)..)).)).))))))..-..((((((((((.....))))))...)))))))).............. ( -31.00) >consensus GCUCUUUACUUCUCGCUUUUUGGGCGCUAUGUAACACAAUAGAUGCGAUGGCGUGAAAACGAACGAGAAUU_UUAUGUUAGGACUGUAUGUCCUGUGCGCGUGAGCUAUGUAUGAAUGUA ((.......((((((..((((.(.(((((((((.(......).))).))))))).))))....)))))).........((((((.....)))))).))((....)).............. (-29.12 = -28.96 + -0.16)

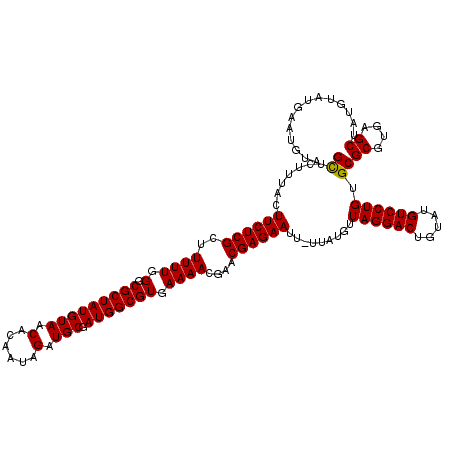
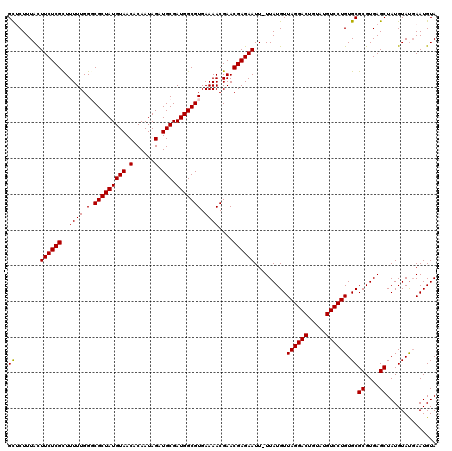
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:45 2006