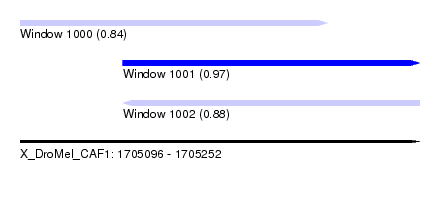
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,705,096 – 1,705,252 |
| Length | 156 |
| Max. P | 0.970154 |

| Location | 1,705,096 – 1,705,216 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -29.90 |
| Energy contribution | -30.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1705096 120 + 22224390 AUCUAAUCGCUGACCACGCACUCGACGCCCAAUGUACUCUGCCCAUAUGUACAUACAUAUGUACGUGAUGGACAUACACAUGUGGGUAUGUGGGUGCUCCUCUCACAUGUGCGAGUAUAC ........((.......))((((((((((((((((....((.((((((((((((....)))))))).)))).))...)))).)))))(((((((.......))))))))).))))).... ( -37.90) >DroSec_CAF1 45047 118 + 1 AUCUAAUCGCUGACUACGCACUCGACGCCCCAUGUACUCUGCCCAUAUGUACAUACAUAUGUACGUGAUGGACAUACCCAUUUGGGUAUGUGGGUGCUCCUCUCACAUGUGCGAGU--AC ............(((.(((((............((((((...((((((((((((....)))))))).))))((((((((....))))))))))))))...........))))))))--.. ( -41.30) >DroSim_CAF1 38967 110 + 1 AUCUAAUCGCUGACCACGCACUCGACGCCCCAUGUACUCUGCCCAUAUGUACAUACAUAUGUACGUGAUGGACAUACCCA--------UGUGGGUGCUCCUCUCACAUGUGCGAGU--AC ................(((((..(((((((((((.....((.((((((((((((....)))))))).)))).))....))--------)).))))).)).........)))))...--.. ( -31.60) >consensus AUCUAAUCGCUGACCACGCACUCGACGCCCCAUGUACUCUGCCCAUAUGUACAUACAUAUGUACGUGAUGGACAUACCCAU_UGGGUAUGUGGGUGCUCCUCUCACAUGUGCGAGU__AC ................(((((............((((((...((((((((((((....)))))))).))))((((((........))))))))))))...........)))))....... (-29.90 = -30.90 + 1.00)

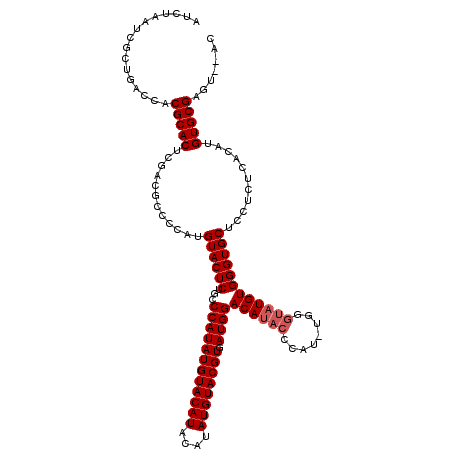
| Location | 1,705,136 – 1,705,252 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 93.35 |
| Mean single sequence MFE | -35.41 |
| Consensus MFE | -31.47 |
| Energy contribution | -32.13 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1705136 116 + 22224390 GCCCAUAUGUACAUACAUAUGUACGUGAUGGACAUACACAUGUGGGUAUGUGGGUGCUCCUCUCACAUGUGCGAGUAUACAAGUAUAGUACAUAUGUAAACCAUAACCCGAUCCAG .....((((....(((((((((((((((.(((((..((((((....))))))..)).)))..)))).(((((..(....)..))))))))))))))))...))))........... ( -34.60) >DroSec_CAF1 45087 114 + 1 GCCCAUAUGUACAUACAUAUGUACGUGAUGGACAUACCCAUUUGGGUAUGUGGGUGCUCCUCUCACAUGUGCGAGU--ACAAGUAUAGUACAUAUGUAAACCAUAACCCGAUCCAG ..((((((((((((....)))))))).))))((((((((....))))))))((((........((.((((((..((--(....))).)))))).)).........))))....... ( -38.43) >DroSim_CAF1 39007 106 + 1 GCCCAUAUGUACAUACAUAUGUACGUGAUGGACAUACCCA--------UGUGGGUGCUCCUCUCACAUGUGCGAGU--ACAAGUAUAGUACAUAUGUAAACCAUAACCCGAUCCAG .....((((....(((((((((((((((.(((..(((((.--------...))))).)))..)))).(((((....--....))))))))))))))))...))))........... ( -33.20) >consensus GCCCAUAUGUACAUACAUAUGUACGUGAUGGACAUACCCAU_UGGGUAUGUGGGUGCUCCUCUCACAUGUGCGAGU__ACAAGUAUAGUACAUAUGUAAACCAUAACCCGAUCCAG .....((((....(((((((((((((((.(((..(((((((........))))))).)))..)))).(((((..(....)..))))))))))))))))...))))........... (-31.47 = -32.13 + 0.67)

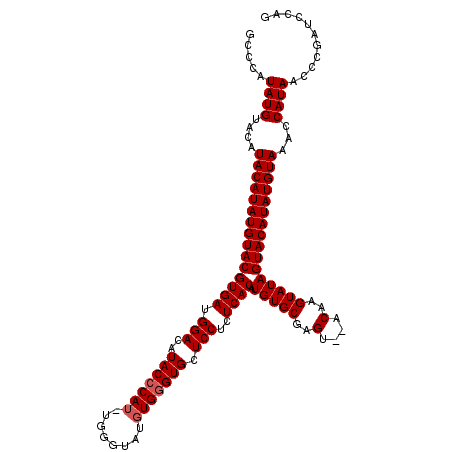
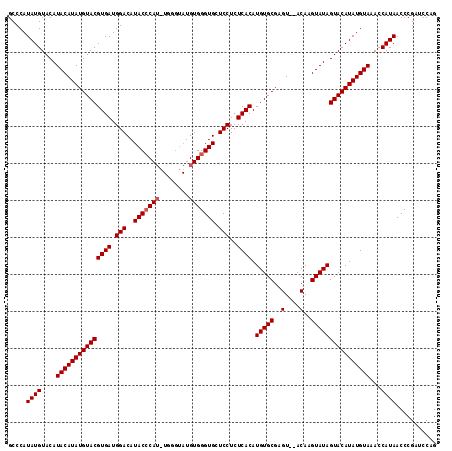
| Location | 1,705,136 – 1,705,252 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 93.35 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -30.27 |
| Energy contribution | -30.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
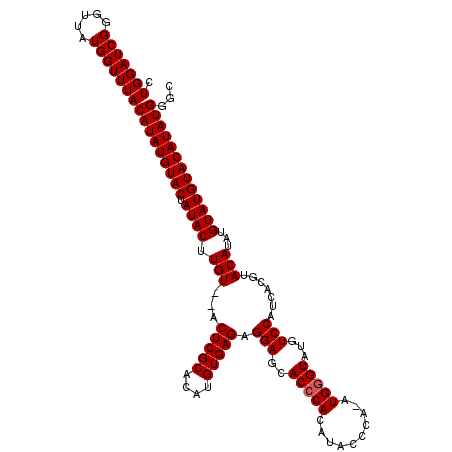
>X_DroMel_CAF1 1705136 116 - 22224390 CUGGAUCGGGUUAUGGUUUACAUAUGUACUAUACUUGUAUACUCGCACAUGUGAGAGGAGCACCCACAUACCCACAUGUGUAUGUCCAUCACGUACAUAUGUAUGUACAUAUGGGC .(((((((.....)))))))(((((((((.((((.(((((.(((((....))))).(((.((..(((((......)))))..))))).....)))))...)))))))))))))... ( -37.60) >DroSec_CAF1 45087 114 - 1 CUGGAUCGGGUUAUGGUUUACAUAUGUACUAUACUUGU--ACUCGCACAUGUGAGAGGAGCACCCACAUACCCAAAUGGGUAUGUCCAUCACGUACAUAUGUAUGUACAUAUGGGC .(((((((.....)))))))(((((((((.((((.(((--((((((....))))((((.....))((((((((....))))))))...))..)))))...)))))))))))))... ( -39.20) >DroSim_CAF1 39007 106 - 1 CUGGAUCGGGUUAUGGUUUACAUAUGUACUAUACUUGU--ACUCGCACAUGUGAGAGGAGCACCCACA--------UGGGUAUGUCCAUCACGUACAUAUGUAUGUACAUAUGGGC .(((((((.....)))))))(((((((((.((((.(((--((((((....))))(((((..((((...--------.))))...))).))..)))))...)))))))))))))... ( -35.80) >consensus CUGGAUCGGGUUAUGGUUUACAUAUGUACUAUACUUGU__ACUCGCACAUGUGAGAGGAGCACCCACAUACCCA_AUGGGUAUGUCCAUCACGUACAUAUGUAUGUACAUAUGGGC .(((((((.....)))))))(((((((((.((((.(((...(((((....))))).(((..(((((..........)))))...))).......)))...)))))))))))))... (-30.27 = -30.60 + 0.33)

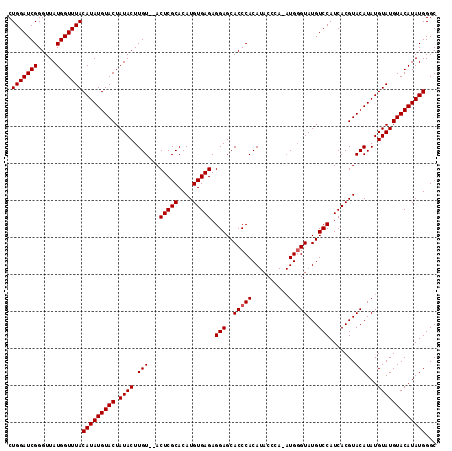
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:34 2006