
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,755,795 – 15,755,976 |
| Length | 181 |
| Max. P | 0.971504 |

| Location | 15,755,795 – 15,755,902 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 98.69 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15755795 107 + 22224390 GCGUCUUGACUUAAUUGCAACAUGCUUUGCGCAUGGCGAAUACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGUCACAACGUGCAGCACUUUCA ..............((((..(((((.....))))))))).........((((..(((((.......)))))..........((((((....)))))).))))..... ( -29.00) >DroSec_CAF1 1976 107 + 1 GCGUCUUGACUUAAUUGCAACAUGCUUUGCGCAUGGCGAAUACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGCCACAACGUGCAGCACUUUCA ..............((((..(((((.....))))))))).........((((..(((((.......)))))..........(((((......))))).))))..... ( -27.90) >DroSim_CAF1 1497 107 + 1 GCGUCUUGACUUAAUUGCAACAUGCUUUGCGCAUGGCGAAUACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGCCACAACGUGCAGCACUUUCA ..............((((..(((((.....))))))))).........((((..(((((.......)))))..........(((((......))))).))))..... ( -27.90) >DroEre_CAF1 1775 107 + 1 GCGUCUUGACUUAAUUGCAACAUGCUUUGCGCAUGGCGAAUACAAACUGUGCCAGCGGCUAUUGAUGCAGCAUUAUAUAAAGCAUGUCACAACGUGCAGCACCUUCA ((((....)).....(((((((((((((((((.(((((...........))))))))(((........)))......)))))))))).......))))))....... ( -27.11) >DroYak_CAF1 1747 107 + 1 GCGUCUUGACUUAAUUGCAACAUGCUUUGCGCAUGGCGAAUACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGUCACAACGUGCAGCACUUUCA ..............((((..(((((.....))))))))).........((((..(((((.......)))))..........((((((....)))))).))))..... ( -29.00) >consensus GCGUCUUGACUUAAUUGCAACAUGCUUUGCGCAUGGCGAAUACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGUCACAACGUGCAGCACUUUCA ..............((((..(((((.....))))))))).........((((..(((((.......)))))..........(((((......))))).))))..... (-27.40 = -27.60 + 0.20)

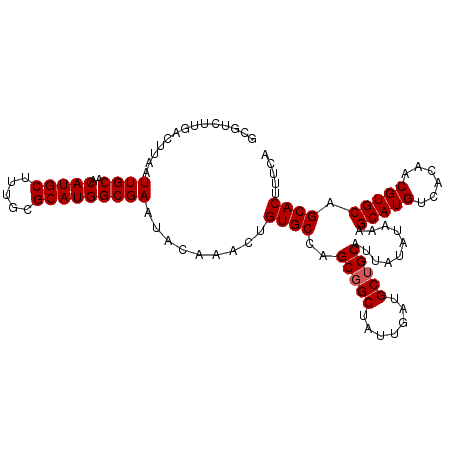
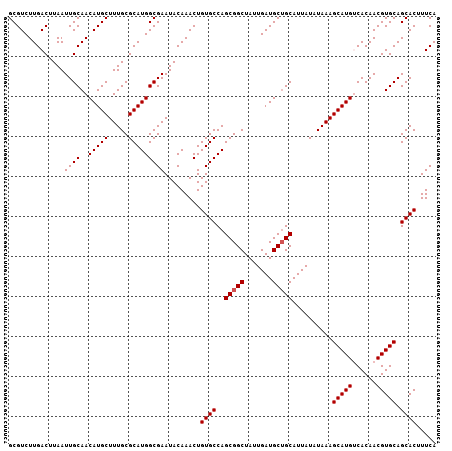
| Location | 15,755,835 – 15,755,942 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 96.54 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15755835 107 + 22224390 UACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGUCACAACGUGCAGCACUUUCAAUACGGACAAGUCCAAAUAGUGAUGAGAAUGUCAGGAUUU ........((((..(((((.......)))))..........((((((....)))))).)))).........(((....)))((((..((((......))))..)))) ( -27.10) >DroSec_CAF1 2016 107 + 1 UACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGCCACAACGUGCAGCACUUUCAAUACGGAAAAGUCCAAAUUGUGAUGAGAAUGUCAGGAUUU ........((((..(((((.......)))))..........(((((......))))).)))).........(((....)))(((((.((((......)))).))))) ( -27.50) >DroSim_CAF1 1537 106 + 1 UACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGCCACAACGUGCAGCACUUUCAAUACGGA-CAGUCCAAAUAGUGAUGAAAAUGUCAGGAUUU .....(((((.((.(((((.......)))))..........(((((......)))))..............)))-))))..((((..((((......))))..)))) ( -24.90) >DroEre_CAF1 1815 107 + 1 UACAAACUGUGCCAGCGGCUAUUGAUGCAGCAUUAUAUAAAGCAUGUCACAACGUGCAGCACCUUCAAUACGGACAAGUCCAAAUAGUGAUGAGAAUGUCAGGAUUU ........((((..((.((.......)).))..........((((((....)))))).)))).........(((....)))((((..((((......))))..)))) ( -22.80) >DroYak_CAF1 1787 107 + 1 UACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGUCACAACGUGCAGCACUUUCAAUACGGACAAGUCCAAAUAGUGAUGAGAAUGUCGGGAUUU .(((....((((..(((((.......)))))..........((((((....)))))).))))(((((.((((((....))).....))).))))).)))........ ( -26.60) >consensus UACAAACUGUGCCAGCGGCUAUUGAUGCUGCAUUAUAUAAAGCAUGUCACAACGUGCAGCACUUUCAAUACGGACAAGUCCAAAUAGUGAUGAGAAUGUCAGGAUUU ........((((..(((((.......)))))..........(((((......))))).)))).........(((....)))((((..((((......))))..)))) (-25.12 = -25.16 + 0.04)


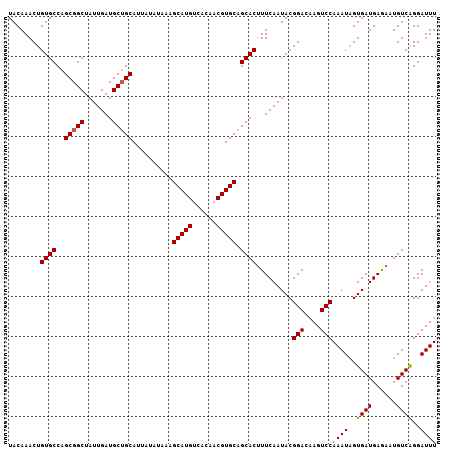
| Location | 15,755,874 – 15,755,976 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.71 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -13.06 |
| Energy contribution | -14.18 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
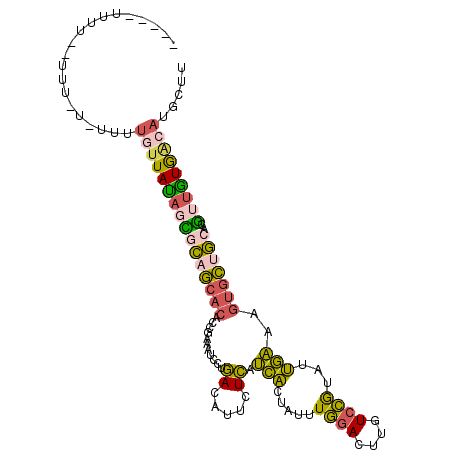
>X_DroMel_CAF1 15755874 102 + 22224390 AAGCAUGUCACAACGUGCAGCACUUUCAAUACGGACAAGUCCAAAUAGUGAUGAGAAUGUCAGGAUUUCGGUGUGCUGCAGUAUAACAAAA-AAAAAAAACAA----- .....(((....((.((((((((..((.....(((....)))((((..((((......))))..)))).)).))))))))))...)))...-...........----- ( -22.20) >DroVir_CAF1 6330 107 + 1 AUACUUUUUAAUAUGUAUACCUAUGAAUAUUUUUAUGCAUUAACUUUAAUUUCACUGUCUGGUGCGACGUAAG-CAAAAACCUUAACAACAAAACAAAAACAACAAAU .............(((.(((..(((((....)))))((((((((............)).))))))...))).)-))................................ ( -8.40) >DroSec_CAF1 2055 101 + 1 AAGCAUGCCACAACGUGCAGCACUUUCAAUACGGAAAAGUCCAAAUUGUGAUGAGAAUGUCAGGAUUUCGCUGUGCUGCGCUAUAACAAAA-AAAAAG-AAAA----- ..............(((((((((.........(((....)))(((((.((((......)))).)))))....)))))))))..........-......-....----- ( -25.40) >DroSim_CAF1 1576 98 + 1 AAGCAUGCCACAACGUGCAGCACUUUCAAUACGGA-CAGUCCAAAUAGUGAUGAAAAUGUCAGGAUUUCGGUGUGCUGCGCUAUAACAAAA---AAAG-AAAA----- ..............(((((((((.......((.((-.(((((......((((......))))))))))).)))))))))))..........---....-....----- ( -22.71) >DroEre_CAF1 1854 93 + 1 AAGCAUGUCACAACGUGCAGCACCUUCAAUACGGACAAGUCCAAAUAGUGAUGAGAAUGUCAGGAUUUCGGUGUGCUGUGCUAUA---AAA-A------AAAA----- ..((((((....))))))(((((.....((((.((..(((((......((((......))))))))))).))))...)))))...---...-.------....----- ( -23.00) >DroYak_CAF1 1826 96 + 1 AAGCAUGUCACAACGUGCAGCACUUUCAAUACGGACAAGUCCAAAUAGUGAUGAGAAUGUCGGGAUUUCGGUGUGCAGCGGUGUAACAAAA-A------AAGG----- ..((((((....)))))).(((((....((((.((..(((((..(((..........)))..))))))).)))).....))))).......-.------....----- ( -21.90) >consensus AAGCAUGUCACAACGUGCAGCACUUUCAAUACGGACAAGUCCAAAUAGUGAUGAGAAUGUCAGGAUUUCGGUGUGCUGCGCUAUAACAAAA_A_AAA__AAAA_____ ..............(((((((((.........(((....)))((((..((((......))))..))))....)))))))))........................... (-13.06 = -14.18 + 1.12)


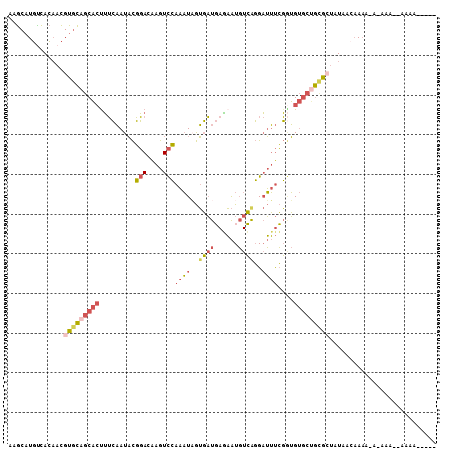
| Location | 15,755,874 – 15,755,976 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 68.71 |
| Mean single sequence MFE | -21.99 |
| Consensus MFE | -15.76 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15755874 102 - 22224390 -----UUGUUUUUUUU-UUUUGUUAUACUGCAGCACACCGAAAUCCUGACAUUCUCAUCACUAUUUGGACUUGUCCGUAUUGAAAGUGCUGCACGUUGUGACAUGCUU -----...........-...((((((((((((((((.....(((.(.((((.((.((........))))..)))).).)))....)))))))).).)))))))..... ( -24.80) >DroVir_CAF1 6330 107 - 1 AUUUGUUGUUUUUGUUUUGUUGUUAAGGUUUUUG-CUUACGUCGCACCAGACAGUGAAAUUAAAGUUAAUGCAUAAAAAUAUUCAUAGGUAUACAUAUUAAAAAGUAU (((((((((((((((...((((.......(((..-((...(((......)))))..))).......))))..)))))))))...)))))).................. ( -11.54) >DroSec_CAF1 2055 101 - 1 -----UUUU-CUUUUU-UUUUGUUAUAGCGCAGCACAGCGAAAUCCUGACAUUCUCAUCACAAUUUGGACUUUUCCGUAUUGAAAGUGCUGCACGUUGUGGCAUGCUU -----....-......-...((((((((((((((((..........(((.....)))...((((.((((....)))).))))...)))))))..)))))))))..... ( -26.70) >DroSim_CAF1 1576 98 - 1 -----UUUU-CUUU---UUUUGUUAUAGCGCAGCACACCGAAAUCCUGACAUUUUCAUCACUAUUUGGACUG-UCCGUAUUGAAAGUGCUGCACGUUGUGGCAUGCUU -----....-....---...((((((((((((((((.....(((.(.((((..((((........)))).))-)).).)))....)))))))..)))))))))..... ( -25.80) >DroEre_CAF1 1854 93 - 1 -----UUUU------U-UUU---UAUAGCACAGCACACCGAAAUCCUGACAUUCUCAUCACUAUUUGGACUUGUCCGUAUUGAAGGUGCUGCACGUUGUGACAUGCUU -----....------.-..(---((((((...(((((((..(((.(.((((.((.((........))))..)))).).)))...)))).)))..)))))))....... ( -20.30) >DroYak_CAF1 1826 96 - 1 -----CCUU------U-UUUUGUUACACCGCUGCACACCGAAAUCCCGACAUUCUCAUCACUAUUUGGACUUGUCCGUAUUGAAAGUGCUGCACGUUGUGACAUGCUU -----....------.-...(((((((.((.(((((((...(((.(.((((.((.((........))))..)))).).)))....))).)))))).)))))))..... ( -22.80) >consensus _____UUUU__UUU_U_UUUUGUUAUAGCGCAGCACACCGAAAUCCUGACAUUCUCAUCACUAUUUGGACUUGUCCGUAUUGAAAGUGCUGCACGUUGUGACAUGCUU ....................((((((((((((((((...........((.....)).(((.....((((....))))...)))..)))))))..)))))))))..... (-15.76 = -15.13 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:33 2006