
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,745,066 – 15,745,185 |
| Length | 119 |
| Max. P | 0.624899 |

| Location | 15,745,066 – 15,745,185 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.54 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -22.27 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15745066 119 + 22224390 AUACUGUGUUUAAUAUGCAUAUGUUCGCGUGUGUGGUGCUCAUGACAUACAUACAUAAAUGCCAACAAAGUCCCAA-AUAUCGCAUUCAUUUGCGAGGGGACUUCGCAGGACGUUAUAAA ...(((((........((((((((....((((((.((....)).))))))..)))))..))).....(((((((..-...(((((......))))).))))))))))))........... ( -36.10) >DroSec_CAF1 4074 120 + 1 AUACUGUGUUUAAUAUGCAUAUGUUCGCGUGUGUGGUGCGCAUGACAUACAUACAUAAAUGCCAAUAAAGUCCGAAAAAAUCGCAUUCAUUUGCGAGGAGACUUCGCAGGACGUUAUAAA .....(((((((.((((..(((((...((((((.....)))))))))))))))..)))))))..((((.(((((((.........)))...((((((....).))))))))).))))... ( -29.50) >DroSim_CAF1 4085 119 + 1 AUACUGUGUUUAAUAUGCAUAUGUUCGCGUGUGUGGUGCGCAUGACAUACAUACAUAAAUGCCAACAAAGUCCCAA-AAAUCGCAUUCAUUUGCGAGGAGACUUCGCAGGACGUUAUAAA ...(((((........((((((((....((((((.((....)).))))))..)))))..))).....(((((((..-...(((((......))))))).))))))))))........... ( -30.30) >DroEre_CAF1 3918 119 + 1 AUACUGUGUUUAAUAUGCAUAUGUUCGCGUGUGUGGUGCGCAUGACACACAUACAUAAAUGCCAACAAAGUCCCAA-AAAUCGCAUCCAUUCGCGAGGGGACUUCGCAGGACGUUAUAAA ...(((((........((((((((....((((((.((....)).))))))..)))))..))).....(((((((..-...((((........)))).))))))))))))........... ( -36.10) >DroYak_CAF1 4010 106 + 1 AUACUGUGUUUAAUAUGCAUAUGUUUGCAUGUGUGGUGCGCAUGACACACAUACAUAAAUGCCAACAAAGUCCCAA-AAAUCGCAUUCGUU-------------CGCAGGACGUUAUAAA ...(((((......((((.(((((.((((((((.....)))))).)).)))))......((....)).........-.....)))).....-------------)))))........... ( -22.50) >consensus AUACUGUGUUUAAUAUGCAUAUGUUCGCGUGUGUGGUGCGCAUGACAUACAUACAUAAAUGCCAACAAAGUCCCAA_AAAUCGCAUUCAUUUGCGAGGAGACUUCGCAGGACGUUAUAAA ....(((((.......)))))((((.(((((((((((((....).))).)))))))....)).))))..((((.......(((((......)))))(....)......))))........ (-22.27 = -22.96 + 0.69)

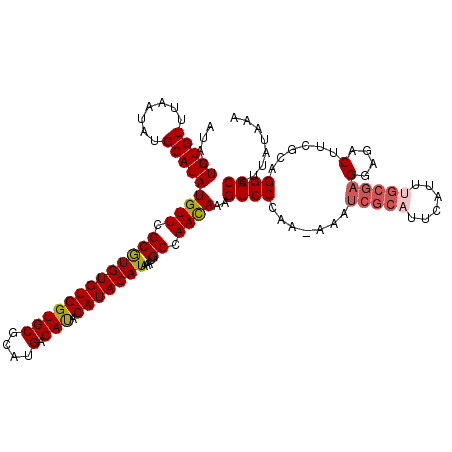
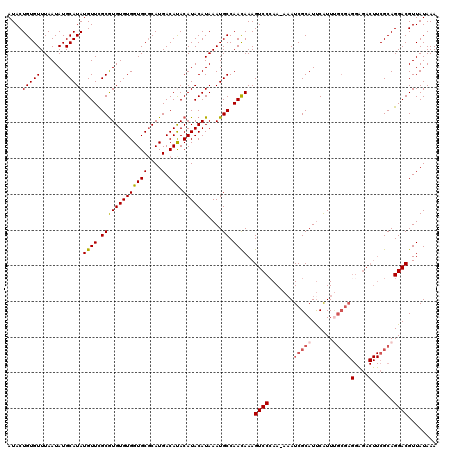
| Location | 15,745,066 – 15,745,185 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.54 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -22.28 |
| Energy contribution | -24.44 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15745066 119 - 22224390 UUUAUAACGUCCUGCGAAGUCCCCUCGCAAAUGAAUGCGAUAU-UUGGGACUUUGUUGGCAUUUAUGUAUGUAUGUCAUGAGCACCACACACGCGAACAUAUGCAUAUUAAACACAGUAU ........(((..((((((((((.(((((......)))))...-..)))))))))).))).((((.((((((((((.....((.........))....))))))))))))))........ ( -32.90) >DroSec_CAF1 4074 120 - 1 UUUAUAACGUCCUGCGAAGUCUCCUCGCAAAUGAAUGCGAUUUUUUCGGACUUUAUUGGCAUUUAUGUAUGUAUGUCAUGCGCACCACACACGCGAACAUAUGCAUAUUAAACACAGUAU ........(((((((((.......))))).........((.....))))))..(((((...((((.((((((((((....(((.........)))...))))))))))))))..))))). ( -26.70) >DroSim_CAF1 4085 119 - 1 UUUAUAACGUCCUGCGAAGUCUCCUCGCAAAUGAAUGCGAUUU-UUGGGACUUUGUUGGCAUUUAUGUAUGUAUGUCAUGCGCACCACACACGCGAACAUAUGCAUAUUAAACACAGUAU ........(((..((((((((((.(((((......)))))...-..)))))))))).))).((((.((((((((((....(((.........)))...))))))))))))))........ ( -32.50) >DroEre_CAF1 3918 119 - 1 UUUAUAACGUCCUGCGAAGUCCCCUCGCGAAUGGAUGCGAUUU-UUGGGACUUUGUUGGCAUUUAUGUAUGUGUGUCAUGCGCACCACACACGCGAACAUAUGCAUAUUAAACACAGUAU ........(((..((((((((((.(((((......)))))...-..)))))))))).)))...(((((((((((....((((.........)))).)))))))))))............. ( -37.00) >DroYak_CAF1 4010 106 - 1 UUUAUAACGUCCUGCG-------------AACGAAUGCGAUUU-UUGGGACUUUGUUGGCAUUUAUGUAUGUGUGUCAUGCGCACCACACAUGCAAACAUAUGCAUAUUAAACACAGUAU ........((((((.(-------------((((....)).)))-.)))))).((((.(.....(((((((((((..((((.(.....).))))...))))))))))).....)))))... ( -25.40) >consensus UUUAUAACGUCCUGCGAAGUCCCCUCGCAAAUGAAUGCGAUUU_UUGGGACUUUGUUGGCAUUUAUGUAUGUAUGUCAUGCGCACCACACACGCGAACAUAUGCAUAUUAAACACAGUAU ........(((..((((((((((.(((((......)))))......)))))))))).))).((((.((((((((((..((((.........))))...))))))))))))))........ (-22.28 = -24.44 + 2.16)

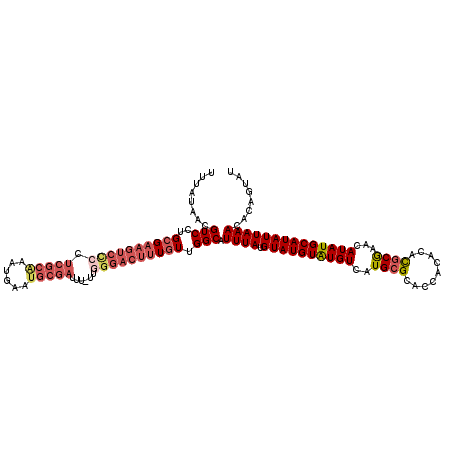
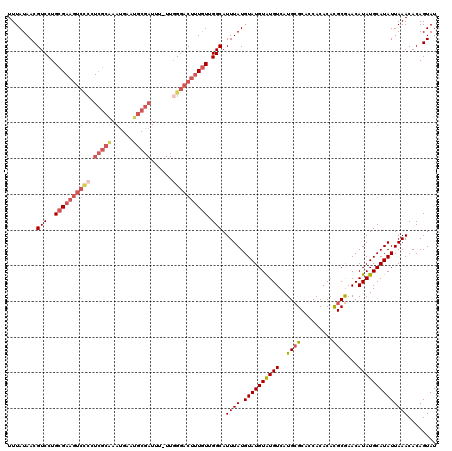
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:27 2006