
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,744,754 – 15,745,026 |
| Length | 272 |
| Max. P | 0.995884 |

| Location | 15,744,754 – 15,744,870 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -36.54 |
| Consensus MFE | -31.36 |
| Energy contribution | -32.60 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15744754 116 - 22224390 CAGCAUCCCUCUGACG-GAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUUUCAAAGUUUUCUCUGAAUCCUUAAGCUUUUUCUGGGAAGCG---AGCUUUUG ..((.((((...((..-((((((((((.(((((.(((.((((..........)))).))).))))))))).(((..(((...)))..)))..)))))))).)))).)).---........ ( -37.20) >DroSec_CAF1 3763 116 - 1 CAGCAUCCCUCUGACG-GAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUUUCAAAGUUUUCUCUGAAUCCUUAAGCUUUUUCUGCGAAGCG---AGCUUUUG .(((.((.((.((.((-((((((((((.(((((.(((.((((..........)))).))).))))))))).(((..(((...)))..)))..)))..))))))).)).)---)))).... ( -33.60) >DroSim_CAF1 3773 116 - 1 CAGCAUCCCUCUGACG-GAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUUUCAAAGUUUUCUCUGAAUCCUUAAGCUUUUUCUGGGAAGCG---AGCUUUUG ..((.((((...((..-((((((((((.(((((.(((.((((..........)))).))).))))))))).(((..(((...)))..)))..)))))))).)))).)).---........ ( -37.20) >DroEre_CAF1 3604 117 - 1 CAGCAUCCCUCUGACG-GUAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUU-AAAAGUUUUCUCUGAAUCCUUAAGCUUUUUCUGGGAAGCA-GAAGCUUUUG ((((...((.(((((.-.........))))).))(((((((.(((......))).))))))))))).(-(((((((...((((..((((............))))..))-)))))))))) ( -37.70) >DroYak_CAF1 3699 118 - 1 CAGCAUCCCUCUGACGCGAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGCGCUGUU-AAAAGUUUUCUCCGAAUCCUUAAGCUU-UUCUGGGAAGCAAGAAGCUUUUG ((((...((.(((((((....))...))))).)).((((((.(((......))).)))))).)))).(-((((((((.((..(..((((.......-....))))..).))))))))))) ( -37.00) >consensus CAGCAUCCCUCUGACG_GAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUUUCAAAGUUUUCUCUGAAUCCUUAAGCUUUUUCUGGGAAGCG___AGCUUUUG ..((.((((...((...((((((((((.(((((.(((.((((..........)))).))).))))))))).(((..(((...)))..)))..)))))))).)))).))............ (-31.36 = -32.60 + 1.24)



| Location | 15,744,791 – 15,744,910 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -34.18 |
| Energy contribution | -34.22 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15744791 119 - 22224390 UUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG-GAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUUUCAAAGUUUUCU (((....)))...................(((((((((......))))).)))).(-((((((((((.(((((.(((.((((..........)))).))).)))))))))...))))))) ( -36.90) >DroSec_CAF1 3800 119 - 1 UUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG-GAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUUUCAAAGUUUUCU (((....)))...................(((((((((......))))).)))).(-((((((((((.(((((.(((.((((..........)))).))).)))))))))...))))))) ( -36.90) >DroSim_CAF1 3810 119 - 1 UUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG-GAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUUUCAAAGUUUUCU (((....)))...................(((((((((......))))).)))).(-((((((((((.(((((.(((.((((..........)))).))).)))))))))...))))))) ( -36.90) >DroEre_CAF1 3643 118 - 1 UUCUUCGGCAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG-GUAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUU-AAAAGUUUUCU ...((((.........))))...........((..(((((((((...((.(((((.-.........))))).))(((((((.(((......))).)))))))))))).-...))))..)) ( -33.10) >DroYak_CAF1 3738 119 - 1 UUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACGCGAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGCGCUGUU-AAAAGUUUUCU ......(..(((.((((((..........(((((((((......))))).))))(((....)))...))((((.(((.((((..........)))).))).)))))))-)...)))..). ( -38.70) >consensus UUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG_GAAAGCGAAGUCGGCGGACGCGAGUGUUCCACUCAACUCUCGUGUGCUGUUUCAAAGUUUUCU ...((((.........)))).........(((((((((......))))).))))...((((((.....(((((.(((.((((..........)))).))).))))).......)))))). (-34.18 = -34.22 + 0.04)


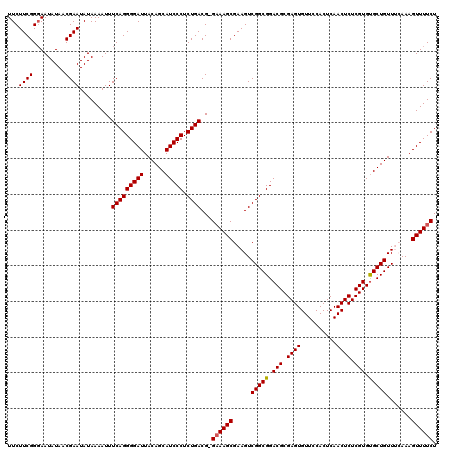
| Location | 15,744,831 – 15,744,950 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -30.24 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15744831 119 - 22224390 CGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG-GAAAGCGAAGUCGGCGGACGCGA ((((.(((((((....)))))))...((.((.((.(((..(((((((.........)))..........(((((((((......))))).)))).)-)))..))))).))))...)))). ( -31.60) >DroSec_CAF1 3840 118 - 1 CGUGAAUGAAAAAUGAUUUUCGUACAGCACGUCUC-UGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG-GAAAGCGAAGUCGGCGGACGCGA .((..(((((((....)))))))...)).(((.((-((((..(((((....((((.....)))).....(((((((((......))))).))))..-.....)))))..)))))).))). ( -33.60) >DroSim_CAF1 3850 119 - 1 CGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG-GAAAGCGAAGUCGGCGGACGCGA ((((.(((((((....)))))))...((.((.((.(((..(((((((.........)))..........(((((((((......))))).)))).)-)))..))))).))))...)))). ( -31.60) >DroEre_CAF1 3682 118 - 1 CGUGAAUGAA-AAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGCAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG-GUAAGCGAAGUCGGCGGACGCGA ..((.(((((-(.....)))))).)).(.(((((.(((.((((((((.........)))).........(((((((((......))))).))))..-......)))).))).))))).). ( -30.20) >DroYak_CAF1 3777 120 - 1 CGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACGCGAAAGCGAAGUCGGCGGACGCGA ..((.(((((((....))))))).)).(.(((((.(((.((((((((.........)))).........(((((((((......))))).)))).((....)))))).))).))))).). ( -35.70) >consensus CGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUACAGCAUCCCUCUGACG_GAAAGCGAAGUCGGCGGACGCGA ..((.(((((((....))))))).)).(.(((((.(((.((((((((.........)))).........(((((((((......))))).)))).........)))).))).))))).). (-30.24 = -30.64 + 0.40)



| Location | 15,744,870 – 15,744,990 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -23.40 |
| Energy contribution | -24.44 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15744870 120 - 22224390 AUCAGCUAAUUGUUCCACACACAUUUGUGUGGGUAAUUUACGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUA ....(((......((((((((....)))))))).....((((.((((((....)))))).)))).)))..((((((((..(((....))).((((.....))))......)))))))).. ( -28.60) >DroSec_CAF1 3879 119 - 1 AUCAGCUAAUUGUUCCGCACACAUUUGUGGUGGGUAUUUACGUGAAUGAAAAAUGAUUUUCGUACAGCACGUCUC-UGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUA ....(((......(((((.(((....))))))))....((((.((((........)))).)))).)))..(((((-(...(((....))).......(((........))).)))))).. ( -24.20) >DroSim_CAF1 3889 120 - 1 AUCAGCUAAUUGUUCCACACACAUUUGUGUGGGUAAUUUACGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUA ....(((......((((((((....)))))))).....((((.((((((....)))))).)))).)))..((((((((..(((....))).((((.....))))......)))))))).. ( -28.60) >DroEre_CAF1 3721 119 - 1 AUCAGCUAAUUGUUCAGCACACAUUUGUGUGGGCAAUUUACGUGAAUGAA-AAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGCAAUAUAACGAAUAUAAAAUUUCAGGGGAUUA ....((((((((((((.((((....)))))))))))))((((.((((((.-..)))))).)))).)))..((((((((((......)))........(((........)))))))))).. ( -26.90) >DroYak_CAF1 3817 120 - 1 AUCAGCUAAUUGUUCCGCACACAUUUGUGUGGGUAAUUUACGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUA ....(((......((((((((....)))))))).....((((.((((((....)))))).)))).)))..((((((((..(((....))).((((.....))))......)))))))).. ( -28.20) >consensus AUCAGCUAAUUGUUCCGCACACAUUUGUGUGGGUAAUUUACGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUCUUCUUCGGGAAUAUAACGAAUAUAAAAUUUCAGGGGAUUA ....(((......((((((((....)))))))).....((((.((((((....)))))).)))).)))..((((((((..(((....))).((((.....))))......)))))))).. (-23.40 = -24.44 + 1.04)


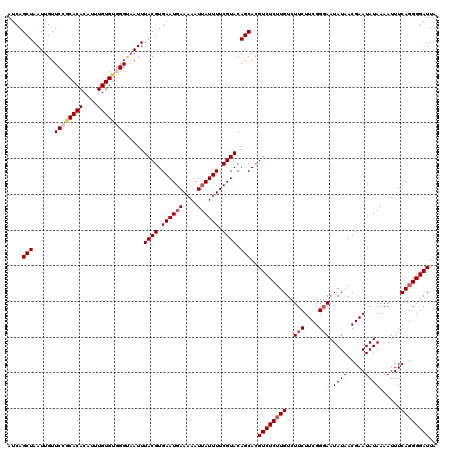
| Location | 15,744,910 – 15,745,026 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -24.56 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15744910 116 - 22224390 GAUUCCAA---U-UAGCAAGUGGCGGGCGAGAACAAUUGAAUCAGCUAAUUGUUCCACACACAUUUGUGUGGGUAAUUUACGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUC ........---.-..(((((.((((.(((.(((((((((.......)))))))))).(((((....))))).......((((.((((((....)))))).))))..)).)))).))))). ( -33.60) >DroSec_CAF1 3919 115 - 1 GAUUCCAA---U-UGGCAAGUGGCGGGCGAGAACAAUUGAAUCAGCUAAUUGUUCCGCACACAUUUGUGGUGGGUAUUUACGUGAAUGAAAAAUGAUUUUCGUACAGCACGUCUC-UGUC ...((((.---.-..(((((((....(((.(((((((((.......))))))))))))...)))))))..)))).....(((((.(((((((....)))))))....)))))...-.... ( -32.80) >DroSim_CAF1 3929 116 - 1 GAUUCCAA---U-UGGCAAGUGGCGGGCGAGAACAAUUGAAUCAGCUAAUUGUUCCACACACAUUUGUGUGGGUAAUUUACGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUC ........---.-.((((((.((((.(((.(((((((((.......)))))))))).(((((....))))).......((((.((((((....)))))).))))..)).)))).)))))) ( -35.20) >DroEre_CAF1 3761 117 - 1 AAUUCCAAGAAUUUCG--AUUUCAAGACAAGAACAAUUGAAUCAGCUAAUUGUUCAGCACACAUUUGUGUGGGCAAUUUACGUGAAUGAA-AAUUAUUUUCGUACAGCACGUCUCUUGUC ........(((.....--..)))..(((((((...............(((((((((.((((....))))))))))))).(((((.(((((-(.....))))))....))))).))))))) ( -26.80) >DroYak_CAF1 3857 113 - 1 AAUUCCAA---UUUAG----CAAACGACAAGAACAAUUGAAUCAGCUAAUUGUUCCGCACACAUUUGUGUGGGUAAUUUACGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUC ........---.....----.....((((((((((((((.......)))))))((((((((....))))))))......(((((.(((((((....)))))))....))))).))))))) ( -28.90) >consensus GAUUCCAA___U_UAGCAAGUGGCGGGCGAGAACAAUUGAAUCAGCUAAUUGUUCCGCACACAUUUGUGUGGGUAAUUUACGUGAAUGAAAAAUUAUUUUCGUACAGCACGUCUCUUGUC .........................((((((((((((((.......)))))))((((((((....))))))))......(((((.(((((((....)))))))....))))).))))))) (-24.56 = -24.92 + 0.36)

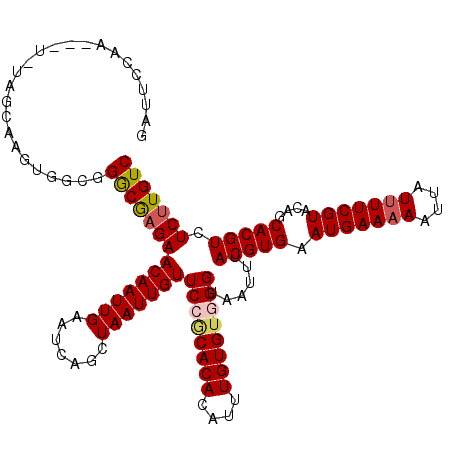
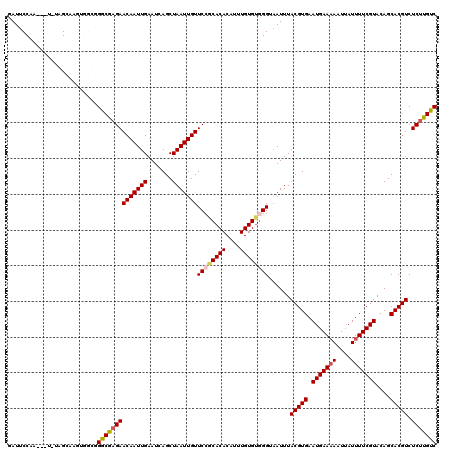
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:25 2006