
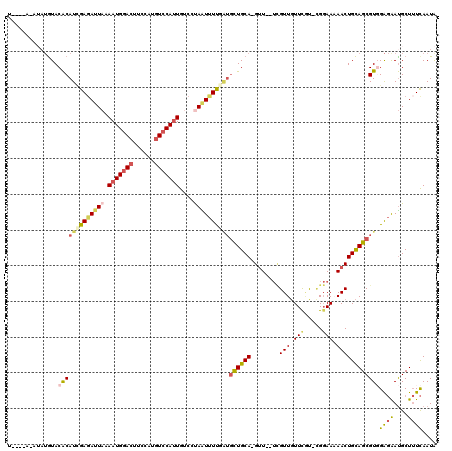
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,744,566 – 15,744,677 |
| Length | 111 |
| Max. P | 0.999793 |

| Location | 15,744,566 – 15,744,677 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -18.18 |
| Energy contribution | -19.70 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.53 |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
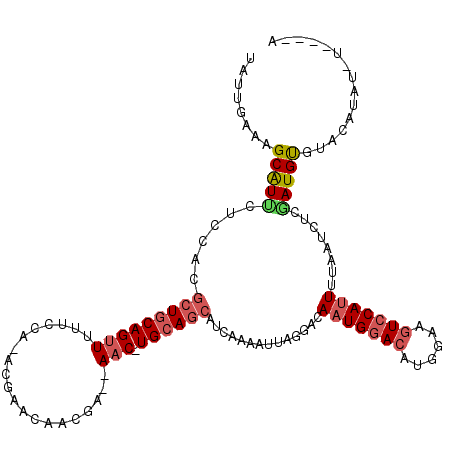
>X_DroMel_CAF1 15744566 111 + 22224390 U--------AUGUACACAUCGAGAUUAAAAUGGACUUCCAUGUCCAUUGUCCUAAUUUCGGUGCUGCA-GUUUUUUGUUGUUUUUUAGGAAAAACUGCAGCGUGGAGAAUGCUUUCAAUG .--------.....(((((((((((((.(((((((......)))))))....))))))))))((((((-(((((((.(........).))))))))))))))))((((....)))).... ( -42.40) >DroSec_CAF1 3578 108 + 1 U--------UUGUACACAUCGAGAUUAAAAUGGACUUCCAUGUCCAUUGUCCUAAUUUUGAUGCUGCA-GUU--UCGUUGUUCGU-UGGAAUAACUGCAGCGUGGAGAAUGCUUUCAAUA .--------.....(((((((((((((.(((((((......)))))))....))))))))))((((((-((.--....(((((..-..))))))))))))))))((((....)))).... ( -37.10) >DroSim_CAF1 3584 112 + 1 U----ACAUAUGUACACAUGGAGAUUAAAAUGGACUUCCAUGUCCAUUGUCCUAAUUUUGAUGCUGCA-GUU--UCGUUGUUCAU-CGGAAUAACUGCAGCGUGGAGAAUGCUUUCAAUA .----.........(((((.(((((((.(((((((......)))))))....))))))).))((((((-(((--...(((.....-)))...))))))))))))((((....)))).... ( -34.10) >DroEre_CAF1 3408 116 + 1 UAUGUAAAUAAGUACGCAUUGAGAAUAAAAUGUAGUUCCAUGUCCAAUGACACAGUCUUGCUUUUGCAAGAU--AAGUUGUUCGC-CG-AAAAACUGCAGCGUUGGCAACGCUAUUAAUA ..........((((.((.(((..(((....(((((((...((.(.((..((...(((((((....)))))))--..))..)).).-))-...)))))))..)))..))).)))))).... ( -24.90) >DroYak_CAF1 3506 116 + 1 AAUGUACAUAUAUACACAUCGACAUUAAAAUGGACUUAGUUGUCCAUUGACCCAGUUUUAAAGCUGCA-GUU--UCGUUCUUCGC-UGGAAAAACUGUAGCGUCGAGGAUGCUUUCAAUA ..((((......))))..(((((.....(((((((......)))))))..............((((((-(((--(..((((....-.)))))))))))))))))))(((....))).... ( -33.20) >consensus U____A_AUAUGUACACAUCGAGAUUAAAAUGGACUUCCAUGUCCAUUGUCCUAAUUUUGAUGCUGCA_GUU__UCGUUGUUCGU_CGGAAAAACUGCAGCGUGGAGAAUGCUUUCAAUA ..............(((((((((((((.(((((((......)))))))....))))))))))((((((........(((.(((.....))).))))))))))))((((....)))).... (-18.18 = -19.70 + 1.52)


| Location | 15,744,566 – 15,744,677 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -13.91 |
| Energy contribution | -14.95 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15744566 111 - 22224390 CAUUGAAAGCAUUCUCCACGCUGCAGUUUUUCCUAAAAAACAACAAAAAAC-UGCAGCACCGAAAUUAGGACAAUGGACAUGGAAGUCCAUUUUAAUCUCGAUGUGUACAU--------A ................(((((((((((((((..............))))))-))))))..(((.(((((...(((((((......)))))))))))).)))..))).....--------. ( -30.94) >DroSec_CAF1 3578 108 - 1 UAUUGAAAGCAUUCUCCACGCUGCAGUUAUUCCA-ACGAACAACGA--AAC-UGCAGCAUCAAAAUUAGGACAAUGGACAUGGAAGUCCAUUUUAAUCUCGAUGUGUACAA--------A ................((((((((((((......-...........--)))-))))))......((..(((.(((((((......)))))))....)))..))))).....--------. ( -26.03) >DroSim_CAF1 3584 112 - 1 UAUUGAAAGCAUUCUCCACGCUGCAGUUAUUCCG-AUGAACAACGA--AAC-UGCAGCAUCAAAAUUAGGACAAUGGACAUGGAAGUCCAUUUUAAUCUCCAUGUGUACAUAUGU----A ........((((....((((((((((((....((-........)).--)))-))))))..........(((.(((((((......)))))))......)))..))).....))))----. ( -28.50) >DroEre_CAF1 3408 116 - 1 UAUUAAUAGCGUUGCCAACGCUGCAGUUUUU-CG-GCGAACAACUU--AUCUUGCAAAAGCAAGACUGUGUCAUUGGACAUGGAACUACAUUUUAUUCUCAAUGCGUACUUAUUUACAUA ........((((((....(((((........-))-)))........--.((((((....))))))(((((((....)))))))................))))))(((......)))... ( -27.10) >DroYak_CAF1 3506 116 - 1 UAUUGAAAGCAUCCUCGACGCUACAGUUUUUCCA-GCGAAGAACGA--AAC-UGCAGCUUUAAAACUGGGUCAAUGGACAACUAAGUCCAUUUUAAUGUCGAUGUGUAUAUAUGUACAUU ..............((((((((.(((((((...(-((..((.....--..)-)...)))..)))))))))).(((((((......))))))).....))))).((((((....)))))). ( -28.10) >consensus UAUUGAAAGCAUUCUCCACGCUGCAGUUUUUCCA_ACGAACAACGA__AAC_UGCAGCAUCAAAAUUAGGACAAUGGACAUGGAAGUCCAUUUUAAUCUCGAUGUGUACAUAU_U____A ........(((((......(((((((((....................))).))))))..............(((((((......)))))))........)))))............... (-13.91 = -14.95 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:21 2006