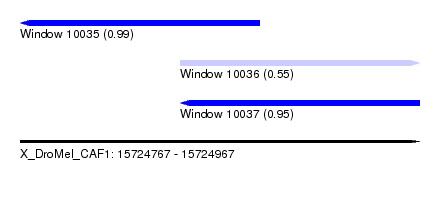
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,724,767 – 15,724,967 |
| Length | 200 |
| Max. P | 0.993868 |

| Location | 15,724,767 – 15,724,887 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -43.09 |
| Consensus MFE | -35.95 |
| Energy contribution | -36.35 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15724767 120 - 22224390 AUUGUUGUGGCUGCAAGUUGUCUUCGAGUGUGGGGUUCCAAACUUUAAGGCGUGCAGCGACUUUUGAUUGCCUGAUGUUCCUGCAUCGGGCGAUGGAUACCAAUGGCUUUUGCUGCCUUU ((((..((.((((((...((((((.((((.(((....))).)))).)))))))))))).))..((.((((((((((((....)))))))))))).))...))))(((.......)))... ( -48.80) >DroSec_CAF1 8198 120 - 1 AUUGUUGUGGCUGGCAGUUGUCUUCGAGUGUGGGGUUCCAAACUUCAAGGCGUGCAUCGACUUCUGAUUGCCUGAUGUUCCUGCAUCGGGCGAUGGAUACCAAUGGCUUUUGCUGCCUUU ............((((((.(((((.((((.(((....))).)))).)))))..((.......(((.((((((((((((....)))))))))))))))........))....))))))... ( -42.96) >DroSim_CAF1 8151 120 - 1 AUUGUUGUGGCUGGCAGUUGUCUUCGAGUGUGGGGUUCCAAACUUCAAGGCGUGCAUCGACUUCUGAUUGCCUGAUGUUCCUGCAUCGGGCGAUGGAUACCAAUGGUUUUUGCUGCCUUU ............((((((.(((((.((((.(((....))).)))).)))))...(((.(...(((.((((((((((((....)))))))))))))))...).)))......))))))... ( -42.30) >DroEre_CAF1 8859 120 - 1 AUUGUUAUGGUUUGCAGUAGUGUUCGAGUGUGGUUUUCCAAAUUUCAGGGCGUGCAGUGACUUUUGAUUGCCUGAUGUACCUGCAUCGGGCGAUGGGUACCAAUGGUUUUUGCUGCCUUU (((((........))))).......(((..(((....)))...)))((((((.((((.((((.(((((((((((((((....))))))))))))......))).)))).)))))))))). ( -38.80) >DroYak_CAF1 7807 120 - 1 AUUGUUGUGACUCGCAGUUGUCUUCAAGUGUGGGGUCCCAAAUUUCAAGGCGUGCAGUGACUUUUGAUUGCCGGAUGCGCCAGCGUCGGGCGAUGGAUACCAAUGGCUUUUGCUGCCCUU ...((((.(.((((((.(((....))).))))))((((((((..(((..(....)..)))..))))((((((.(((((....))))).)))))))))).)))))(((.......)))... ( -42.60) >consensus AUUGUUGUGGCUGGCAGUUGUCUUCGAGUGUGGGGUUCCAAACUUCAAGGCGUGCAGCGACUUUUGAUUGCCUGAUGUUCCUGCAUCGGGCGAUGGAUACCAAUGGCUUUUGCUGCCUUU ............((((((.(((((.((((.(((....))).)))).)))))..((........((.((((((((((((....)))))))))))).))........))....))))))... (-35.95 = -36.35 + 0.40)

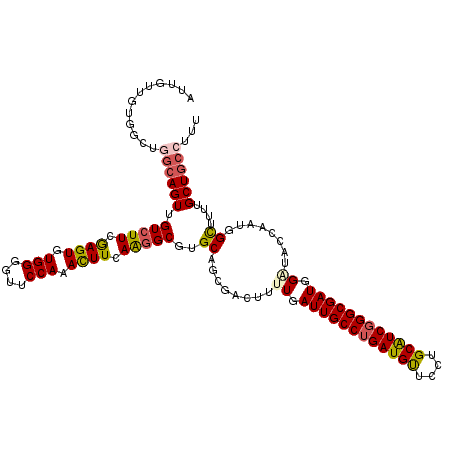
| Location | 15,724,847 – 15,724,967 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -28.06 |
| Energy contribution | -27.50 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15724847 120 + 22224390 UGGAACCCCACACUCGAAGACAACUUGCAGCCACAACAAUCGACGCAUGUCUGUUCUGAACAAGUUGUUCAUGACGAACAUCACUGAUAUUCUGGCCACCGGCGCAGUUGCCGAUUCCAU (((((........................((((.....((((..(.((((.((((.(((((.....))))).)))).)))))..))))....))))...(((((....))))).))))). ( -28.50) >DroSec_CAF1 8278 120 + 1 UGGAACCCCACACUCGAAGACAACUGCCAGCCACAACAAUCGACGCAUGUCUGUUCUGAACAAGUUGUUCAUGACGAACAUCACUGAUAUUCUGGCCACCGGCGCAGUUGCCGAUUCCAU (((((........(((..(.((((((((.((((.....((((..(.((((.((((.(((((.....))))).)))).)))))..))))....)))).....).)))))))))))))))). ( -30.60) >DroSim_CAF1 8231 120 + 1 UGGAACCCCACACUCGAAGACAACUGCCAGCCACAACAAUCGACGCAUGUCUGUUCUGAACAAGUUGUUCAUGACGAACAUCACUGAUAUUCUGGCCACCGGCGCAGUUGCCGAUUCCAU (((((........(((..(.((((((((.((((.....((((..(.((((.((((.(((((.....))))).)))).)))))..))))....)))).....).)))))))))))))))). ( -30.60) >DroEre_CAF1 8939 120 + 1 UGGAAAACCACACUCGAACACUACUGCAAACCAUAACAAUCGACGCAUGUCCGUGCUGAACAAGUUGUUCAUGACGAACAUCACCGAUAUUCUGGCCACCGGCGCAGUUGCCGAUUCCAU (((((........(((......(((((...........((((..(.((((.(((..(((((.....)))))..))).)))))..))))......(((...))))))))...)))))))). ( -28.10) >DroYak_CAF1 7887 120 + 1 UGGGACCCCACACUUGAAGACAACUGCGAGUCACAACAAUCGACGCAUGUCCGUUCUGAACAAGUUGUUCAUGACGAACAUCACCGAUAUUCUGGCCACCGGCGCAGUGGCCGAUUCUAU (((....))).................(((((......((((..(.((((.((((.(((((.....))))).)))).)))))..)))).....((((((.......)))))))))))... ( -35.70) >consensus UGGAACCCCACACUCGAAGACAACUGCCAGCCACAACAAUCGACGCAUGUCUGUUCUGAACAAGUUGUUCAUGACGAACAUCACUGAUAUUCUGGCCACCGGCGCAGUUGCCGAUUCCAU (((((........................((((.....((((..(.((((.((((.(((((.....))))).)))).)))))..))))....))))...((((......)))).))))). (-28.06 = -27.50 + -0.56)

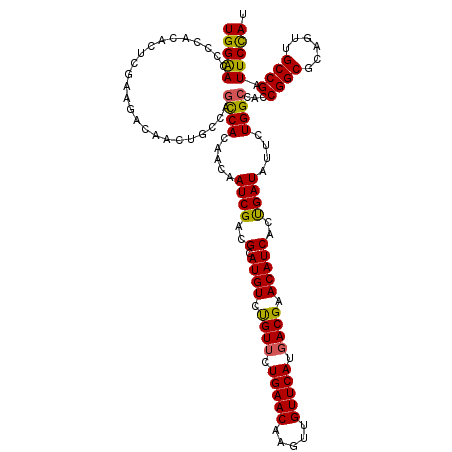
| Location | 15,724,847 – 15,724,967 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -44.44 |
| Consensus MFE | -34.96 |
| Energy contribution | -37.04 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15724847 120 - 22224390 AUGGAAUCGGCAACUGCGCCGGUGGCCAGAAUAUCAGUGAUGUUCGUCAUGAACAACUUGUUCAGAACAGACAUGCGUCGAUUGUUGUGGCUGCAAGUUGUCUUCGAGUGUGGGGUUCCA .((((((((((((((......(..((((.((((((..((((((..((..(((((.....)))))..))..)))).))..)).)))).))))..).))))))).(((....)))))))))) ( -43.50) >DroSec_CAF1 8278 120 - 1 AUGGAAUCGGCAACUGCGCCGGUGGCCAGAAUAUCAGUGAUGUUCGUCAUGAACAACUUGUUCAGAACAGACAUGCGUCGAUUGUUGUGGCUGGCAGUUGUCUUCGAGUGUGGGGUUCCA .((((((((((((((((.(....(((((.((((((..((((((..((..(((((.....)))))..))..)))).))..)).)))).))))))))))))))).(((....)))))))))) ( -45.30) >DroSim_CAF1 8231 120 - 1 AUGGAAUCGGCAACUGCGCCGGUGGCCAGAAUAUCAGUGAUGUUCGUCAUGAACAACUUGUUCAGAACAGACAUGCGUCGAUUGUUGUGGCUGGCAGUUGUCUUCGAGUGUGGGGUUCCA .((((((((((((((((.(....(((((.((((((..((((((..((..(((((.....)))))..))..)))).))..)).)))).))))))))))))))).(((....)))))))))) ( -45.30) >DroEre_CAF1 8939 120 - 1 AUGGAAUCGGCAACUGCGCCGGUGGCCAGAAUAUCGGUGAUGUUCGUCAUGAACAACUUGUUCAGCACGGACAUGCGUCGAUUGUUAUGGUUUGCAGUAGUGUUCGAGUGUGGUUUUCCA .(((((.(.(((...((((..(..((((.(((((((..(((((((((..(((((.....)))))..)))))))).)..))).)))).))).)..)....)))).....))).)..))))) ( -40.50) >DroYak_CAF1 7887 120 - 1 AUAGAAUCGGCCACUGCGCCGGUGGCCAGAAUAUCGGUGAUGUUCGUCAUGAACAACUUGUUCAGAACGGACAUGCGUCGAUUGUUGUGACUCGCAGUUGUCUUCAAGUGUGGGGUCCCA ........((((((((...))))))))(.(((((((..(((((((((..(((((.....)))))..)))))))).)..))).)))).)..((((((.(((....))).))))))...... ( -47.60) >consensus AUGGAAUCGGCAACUGCGCCGGUGGCCAGAAUAUCAGUGAUGUUCGUCAUGAACAACUUGUUCAGAACAGACAUGCGUCGAUUGUUGUGGCUGGCAGUUGUCUUCGAGUGUGGGGUUCCA .((((((((((((((((.....((((((.((((....((((((..(((.(((((.....))))).....)))..))))))..)))).))))))))))))))).(((....)))))))))) (-34.96 = -37.04 + 2.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:04 2006