
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,723,836 – 15,724,035 |
| Length | 199 |
| Max. P | 0.996829 |

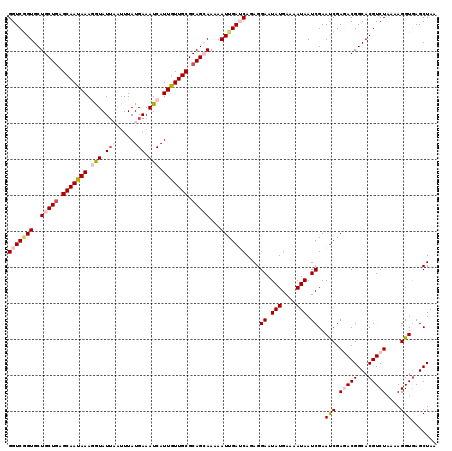
| Location | 15,723,836 – 15,723,955 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -22.30 |
| Energy contribution | -24.22 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15723836 119 - 22224390 GGUCGGUGCUGCUGAGCAAUAAAGGUAUUAAUUUAUGAAAUCAUUGUUGCGCAGCAAAAAUUGAUCAGAGGAAUAUGAAAAUAAUCGAAUCGAGACGGCACGUCUAAAAGGUGAGCUAA (((((((..(((((.(((((((.(((.(((.....))).))).))))))).)))))...)))))))....((.(((....))).))..(((.(((((...)))))....)))....... ( -31.40) >DroSec_CAF1 7378 115 - 1 GGUCGGUGCUGCUGAGCAAUAAAGGUAU----UUAUGAAAUCAUUGUUGCGCAGCAAAAAUUGAUCAGAGGAAUAUGAAAAUAAUCGAACCGAGACGGCACGUCUAAAAGGUAAGCUAA (((((((..(((((.(((((((.(((..----.......))).))))))).)))))...)))))))....((.(((....))).))..(((.(((((...)))))....)))....... ( -32.80) >DroSim_CAF1 7328 118 - 1 GGUC-GUGCUGCUGAGCAAUAAAGGUAUUAAUUUAUGAAAUCAUUGUUGCGCAGCAAAAAUUGAUCAGAGGAAUAUGAAAAUAAUCGAAUCGAGACGGCACGUCUAAAAGGUGAGCUAA ((.(-(((((((((.(((((((.(((.(((.....))).))).))))))).)))).....(((((..((....(((....))).))..)))))...)))))).)).............. ( -32.00) >DroEre_CAF1 7715 119 - 1 GGUCAGUGCUGCUGAGCAACAAAUAUAUUCAUUUAUGAAAUCAUUGUUGCGCAGAAAGAAUUGAACACAGGAAUAUGAAAAUAAUCGAAUCGAAACGGCACGUCUAAAAGGUGAGCUAA (.((((((((((((.(((((((.....((((....))))....))))))).)))................((.(((....))).)).........)))))).((.....))))).)... ( -25.00) >DroYak_CAF1 6827 119 - 1 GGUCGGUGCUGCUCGGCAAUAAAAAUAUUAAUUUAUGAAAUCAUUGUUGCGCAGAAAGAAUUGAACACAGGAAUAUGAGAAUAAUCGAAUCGAAACGGCACGUCUAAAAGGUGAGCUAA ..(((((.((((.(((((((....(((......)))......))))))).))))................((.(((....))).))..)))))...(((.(..(.....)..).))).. ( -20.50) >consensus GGUCGGUGCUGCUGAGCAAUAAAGGUAUUAAUUUAUGAAAUCAUUGUUGCGCAGCAAAAAUUGAUCAGAGGAAUAUGAAAAUAAUCGAAUCGAGACGGCACGUCUAAAAGGUGAGCUAA (((((((..(((((.(((((((.(((.(((.....))).))).))))))).)))))...)))))))....((.(((....))).))..(((.(((((...)))))....)))....... (-22.30 = -24.22 + 1.92)


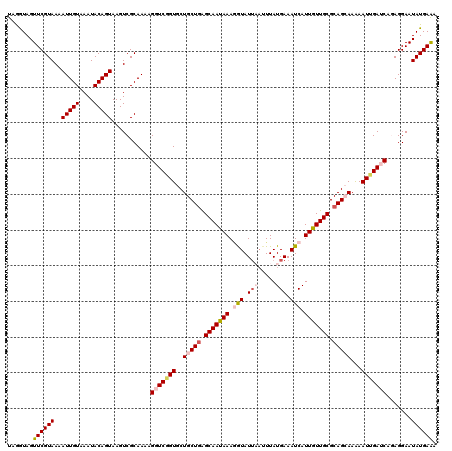
| Location | 15,723,875 – 15,723,995 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -22.92 |
| Energy contribution | -24.44 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15723875 120 - 22224390 UAGGUAGUUCGUAAAAUUGUAAAUACAGUAAGUCGCAAAAGGUCGGUGCUGCUGAGCAAUAAAGGUAUUAAUUUAUGAAAUCAUUGUUGCGCAGCAAAAAUUGAUCAGAGGAAUAUGAAA .......((((((..(((((....)))))...........(((((((..(((((.(((((((.(((.(((.....))).))).))))))).)))))...))))))).......)))))). ( -30.10) >DroSec_CAF1 7417 116 - 1 UAAGUAGUUCGUAAAAUUGUAAAUACAGUAAGUCGCAAAAGGUCGGUGCUGCUGAGCAAUAAAGGUAU----UUAUGAAAUCAUUGUUGCGCAGCAAAAAUUGAUCAGAGGAAUAUGAAA .......((((((..(((((....)))))...........(((((((..(((((.(((((((.(((..----.......))).))))))).)))))...))))))).......)))))). ( -28.90) >DroSim_CAF1 7367 119 - 1 UAAGUAGUUCGUAAAAUUGUAAAUACAGUAAGUCGCAAAAGGUC-GUGCUGCUGAGCAAUAAAGGUAUUAAUUUAUGAAAUCAUUGUUGCGCAGCAAAAAUUGAUCAGAGGAAUAUGAAA .......((((((..(((((....)))))...........((((-(...(((((.(((((((.(((.(((.....))).))).))))))).))))).....))))).......)))))). ( -26.80) >DroEre_CAF1 7754 120 - 1 UAGAUAGUUCGUAAAAUUGUAAAUACAGUAAGUCGCAAAAGGUCAGUGCUGCUGAGCAACAAAUAUAUUCAUUUAUGAAAUCAUUGUUGCGCAGAAAGAAUUGAACACAGGAAUAUGAAA .......((((((...((((.....(((((....((.....))...)))))(((.(((((((.....((((....))))....))))))).)))............))))...)))))). ( -23.30) >DroYak_CAF1 6866 120 - 1 UUGGUAGUUCGUAAAAUUGUAAAUACAGUAAGUCGCAAAAGGUCGGUGCUGCUCGGCAAUAAAAAUAUUAAUUUAUGAAAUCAUUGUUGCGCAGAAAGAAUUGAACACAGGAAUAUGAGA .......((((((..(((((....)))))...........(.(((((.((((.(((((((....(((......)))......))))))).)))).....))))).).......)))))). ( -19.70) >consensus UAGGUAGUUCGUAAAAUUGUAAAUACAGUAAGUCGCAAAAGGUCGGUGCUGCUGAGCAAUAAAGGUAUUAAUUUAUGAAAUCAUUGUUGCGCAGCAAAAAUUGAUCAGAGGAAUAUGAAA .......((((((..(((((....)))))...........(((((((..(((((.(((((((.(((.(((.....))).))).))))))).)))))...))))))).......)))))). (-22.92 = -24.44 + 1.52)

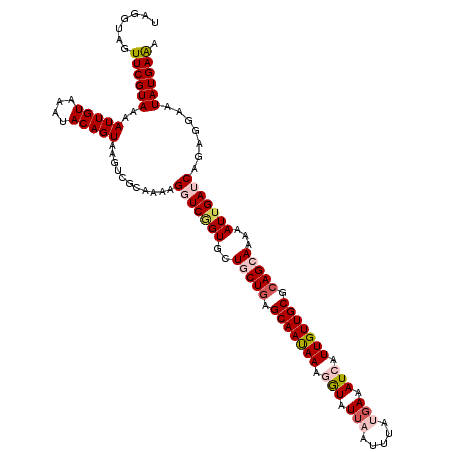
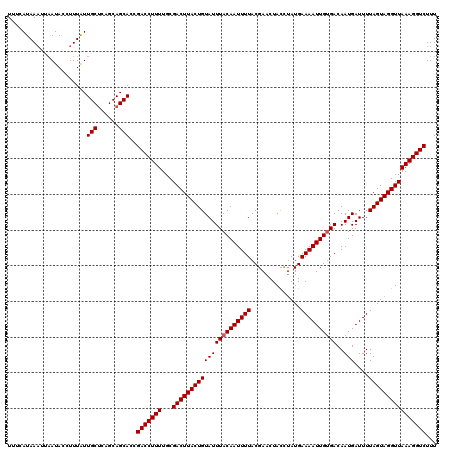
| Location | 15,723,915 – 15,724,035 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -23.19 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15723915 120 + 22224390 UUUCAUAAAUUAAUACCUUUAUUGCUCAGCAGCACCGACCUUUUGCGACUUACUGUAUUUACAAUUUUACGAACUACCUAUGAAAAUUGUGACAAUGAUUUUAGUAGGUUAAAGGUCUUU ......................((((....))))..(((((((...((((((((((((((((((((((...(......)...))))))))))..)))....))))))))))))))))... ( -23.10) >DroSec_CAF1 7457 116 + 1 UUUCAUAA----AUACCUUUAUUGCUCAGCAGCACCGACCUUUUGCGACUUACUGUAUUUACAAUUUUACGAACUACUUAUGAAAAUUGUGACAAUGAUUUUAGUAGGUUAAAGGUCUUU ........----..........((((....))))..(((((((...((((((((((((((((((((((...((....))...))))))))))..)))....))))))))))))))))... ( -23.50) >DroSim_CAF1 7407 119 + 1 UUUCAUAAAUUAAUACCUUUAUUGCUCAGCAGCAC-GACCUUUUGCGACUUACUGUAUUUACAAUUUUACGAACUACUUAUGAAAAUUGUGACAAUGAUUUUAGUAGGUUAAAGGUCUUU ......................((((....)))).-(((((((...((((((((((((((((((((((...((....))...))))))))))..)))....))))))))))))))))... ( -23.50) >DroEre_CAF1 7794 120 + 1 UUUCAUAAAUGAAUAUAUUUGUUGCUCAGCAGCACUGACCUUUUGCGACUUACUGUAUUUACAAUUUUACGAACUAUCUAUGAAAAUUGCGACAAUGAUUUUAGUAGGUUAAAGGUCUUU .((((....))))......((((((...))))))..(((((((...(((((((((...((((((((((..((....))....)))))))......)))...))))))))))))))))... ( -23.90) >DroYak_CAF1 6906 120 + 1 UUUCAUAAAUUAAUAUUUUUAUUGCCGAGCAGCACCGACCUUUUGCGACUUACUGUAUUUACAAUUUUACGAACUACCAAUGAAAAUUGUGACAAUGAUUUUAGUAGGUUAAAGGUCUUU ......................(((......)))..(((((((...((((((((((((((((((((((..............))))))))))..)))....))))))))))))))))... ( -21.94) >consensus UUUCAUAAAUUAAUACCUUUAUUGCUCAGCAGCACCGACCUUUUGCGACUUACUGUAUUUACAAUUUUACGAACUACCUAUGAAAAUUGUGACAAUGAUUUUAGUAGGUUAAAGGUCUUU ......................(((......)))..(((((((...((((((((((((((((((((((..............))))))))))..)))....))))))))))))))))... (-21.54 = -21.74 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:01 2006