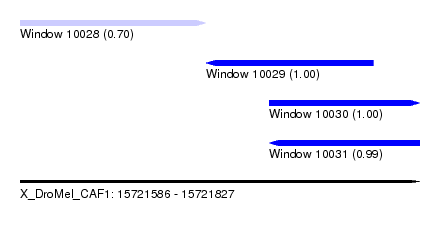
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,721,586 – 15,721,827 |
| Length | 241 |
| Max. P | 0.999643 |

| Location | 15,721,586 – 15,721,698 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 99.29 |
| Mean single sequence MFE | -17.68 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15721586 112 + 22224390 CUCCUCCUUAAAUAAUUUUAUGAUUUCCGUUUGUUACAUUUGUAUGAAUAUACGACUAUAAUAUAUGGAACAAGAUCACAUCACAACGUGAAUUAUAGUAACAACUAUAGCU ........................((((((.(((((.(.((((((....)))))).).))))).)))))).....((((........)))).(((((((....))))))).. ( -18.20) >DroSec_CAF1 5100 112 + 1 CUCCUCCUUAAAUAAUUUUAUGAUUUCCGUUUGUUACAUUUGUAUGAAUAUACGACUAUAAUAUAUGGAACAAGAUCACAUCACAACGUGAAUUAUAGUAACAACUAUAGCU ........................((((((.(((((.(.((((((....)))))).).))))).)))))).....((((........)))).(((((((....))))))).. ( -18.20) >DroSim_CAF1 4780 112 + 1 CUCCUCCUUAAAUAAUUUUAUGAUUUCCGUUUGUUACAUUUGUAUGAAUAUACGACUAUAAUAUAUGGAACAAGAUCACAUCACAACGUGAAUUAUAGUAAUAACUAUAGCU ........................((((((.(((((.(.((((((....)))))).).))))).)))))).....((((........)))).(((((((....))))))).. ( -18.20) >DroEre_CAF1 5484 112 + 1 CUCCUCCUUAAAUAAUUUUAUGAUUUCCGUUUGAUACAUUUGUAUGAAUAUACGACUAUAAUAUAUGGAACAAGAUCACAUCACAACGUGAAUUAUAGUAACAACUAUAGCU .................((((((((..((((((((....((((((....))))))(((((...)))))...........)))).))))..)))))))).............. ( -15.60) >DroYak_CAF1 4591 112 + 1 CUCCUCCUUAAAUAAUUUUAUGAUUUCCGUUUGUUACAUUUGUAUGAAUAUACGACUAUAAUAUAUGGAACAAGAUCACAUCACAACGUGAAUUAUAGUAACAACUAUAGCU ........................((((((.(((((.(.((((((....)))))).).))))).)))))).....((((........)))).(((((((....))))))).. ( -18.20) >consensus CUCCUCCUUAAAUAAUUUUAUGAUUUCCGUUUGUUACAUUUGUAUGAAUAUACGACUAUAAUAUAUGGAACAAGAUCACAUCACAACGUGAAUUAUAGUAACAACUAUAGCU ........................((((((.(((((.(.((((((....)))))).).))))).)))))).....((((........)))).(((((((....))))))).. (-17.54 = -17.74 + 0.20)

| Location | 15,721,698 – 15,721,799 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15721698 101 - 22224390 CGUACAAAUCAAGGCAUUAAAUUUAUCGUGAACAUUUCAAAAGAAUUUAAAACAGAUAAAAUCGUUUUUGAAUGUUAUUCUUAGAGUGCAGCCUUGACAAA ........(((((((.((((((((....(((.....)))...))))))))((((..(((((....)))))..))))..............))))))).... ( -19.30) >DroSec_CAF1 5212 101 - 1 CGUACAAAUCAAGGCAUUAAAUUUAUCGUGAACAUUUCAGAAGAAUUUAAAGCAGAUAAAAUCGUUUUUGAAUGUUAUUCUUAGAGUGCAGCCUUGACAAA ........(((((((.((((((((.((.(((.....))))).))))))))((((..(((((....)))))..))))..............))))))).... ( -21.30) >DroSim_CAF1 4892 101 - 1 CGUACAAAUCAAGGCAUUAAAUUUAUCGUGAACAUUUCAGAAGAAUUUAAAACAGAUAAAAUCGUUUUUGAAUGUUAUUCUUAGAGUGCAGCCUUGACAAA ........(((((((.((((((((.((.(((.....))))).))))))))((((..(((((....)))))..))))..............))))))).... ( -21.30) >DroEre_CAF1 5596 98 - 1 CAUACAAAUCAAGGCAUUAAAUUUAUCAUGAACAUUUCAGAAGAAUUUAAAACAAAUACAAUCGUUUUUGAAUGUUAU---UAGAGUGCAGCCUUGACAAA ........(((((((.((((((((.((.(((.....))))).))))))))..((((.((....)).))))........---.........))))))).... ( -20.00) >DroYak_CAF1 4703 101 - 1 CAUACAAAUCAAGGCAUUAAAUUUAUCAUGAACAUUUCAAAAGAAUUUAAAACAGAUGCAAUCGUUUUUGAAUGUUAUUCUUAGAGUGCAGCCUUGACAAA ........(((((((.((((((((....(((.....)))...))))))))......((((.((......((((...))))...)).))))))))))).... ( -19.80) >consensus CGUACAAAUCAAGGCAUUAAAUUUAUCGUGAACAUUUCAGAAGAAUUUAAAACAGAUAAAAUCGUUUUUGAAUGUUAUUCUUAGAGUGCAGCCUUGACAAA ........(((((((......((((.....(((((((.((((((.((((.......)))).)).)))).))))))).....)))).....))))))).... (-18.18 = -18.18 + -0.00)



| Location | 15,721,736 – 15,721,827 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -19.10 |
| Energy contribution | -18.86 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15721736 91 + 22224390 GAUUUUAUCUGUUUUAAAUUCUUUUGAAAUGUUCACGAUAAAUUUAAUGCCUUGAUUUGUACGGUUCAAAUUGGGGCGUGUUCUUUAAAUG ...((((((.((((((((....))))))))(....)))))))....((((((..(((((.......)))))..))))))............ ( -20.60) >DroSec_CAF1 5250 91 + 1 GAUUUUAUCUGCUUUAAAUUCUUCUGAAAUGUUCACGAUAAAUUUAAUGCCUUGAUUUGUACGGUUCAAAUUGGGGCGUGUUCUUUAAAUG .((((.....((..((((((..(((((.....))).))..))))))((((((..(((((.......)))))..)))))))).....)))). ( -20.00) >DroSim_CAF1 4930 91 + 1 GAUUUUAUCUGUUUUAAAUUCUUCUGAAAUGUUCACGAUAAAUUUAAUGCCUUGAUUUGUACGGUUCAAAUUGGGGCGUGUUUUUUAAAUG ..............((((((..(((((.....))).))..))))))((((((..(((((.......)))))..))))))............ ( -19.90) >DroEre_CAF1 5631 91 + 1 GAUUGUAUUUGUUUUAAAUUCUUCUGAAAUGUUCAUGAUAAAUUUAAUGCCUUGAUUUGUAUGGUUCAAAUUGGGGCGUAUUUUUUAAAUG .....((((((...((((((..(((((.....))).))..))))))((((((..(((((.......)))))..))))))......)))))) ( -20.20) >DroYak_CAF1 4741 91 + 1 GAUUGCAUCUGUUUUAAAUUCUUUUGAAAUGUUCAUGAUAAAUUUAAUGCCUUGAUUUGUAUGGUUCAAAUUGGGGCGUGUUCUUUAAAUG ..........((((((((....))))))))...(((..((((....((((((..(((((.......)))))..))))))....)))).))) ( -19.60) >consensus GAUUUUAUCUGUUUUAAAUUCUUCUGAAAUGUUCACGAUAAAUUUAAUGCCUUGAUUUGUACGGUUCAAAUUGGGGCGUGUUCUUUAAAUG ..............((((((..(((((.....))).))..))))))((((((..(((((.......)))))..))))))............ (-19.10 = -18.86 + -0.24)


| Location | 15,721,736 – 15,721,827 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -11.72 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.50 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15721736 91 - 22224390 CAUUUAAAGAACACGCCCCAAUUUGAACCGUACAAAUCAAGGCAUUAAAUUUAUCGUGAACAUUUCAAAAGAAUUUAAAACAGAUAAAAUC ..............(((...(((((.......)))))...)))......((((((((......(((....)))......)).))))))... ( -11.10) >DroSec_CAF1 5250 91 - 1 CAUUUAAAGAACACGCCCCAAUUUGAACCGUACAAAUCAAGGCAUUAAAUUUAUCGUGAACAUUUCAGAAGAAUUUAAAGCAGAUAAAAUC ..............(((...(((((.......)))))...))).((((((((.((.(((.....))))).))))))))............. ( -12.30) >DroSim_CAF1 4930 91 - 1 CAUUUAAAAAACACGCCCCAAUUUGAACCGUACAAAUCAAGGCAUUAAAUUUAUCGUGAACAUUUCAGAAGAAUUUAAAACAGAUAAAAUC ..............(((...(((((.......)))))...))).((((((((.((.(((.....))))).))))))))............. ( -12.30) >DroEre_CAF1 5631 91 - 1 CAUUUAAAAAAUACGCCCCAAUUUGAACCAUACAAAUCAAGGCAUUAAAUUUAUCAUGAACAUUUCAGAAGAAUUUAAAACAAAUACAAUC ..............(((...(((((.......)))))...))).((((((((.((.(((.....))))).))))))))............. ( -12.30) >DroYak_CAF1 4741 91 - 1 CAUUUAAAGAACACGCCCCAAUUUGAACCAUACAAAUCAAGGCAUUAAAUUUAUCAUGAACAUUUCAAAAGAAUUUAAAACAGAUGCAAUC (((((.........(((...(((((.......)))))...))).((((((((....(((.....)))...))))))))...)))))..... ( -10.60) >consensus CAUUUAAAGAACACGCCCCAAUUUGAACCGUACAAAUCAAGGCAUUAAAUUUAUCGUGAACAUUUCAGAAGAAUUUAAAACAGAUAAAAUC ..............(((...(((((.......)))))...))).((((((((.((.(((.....))))).))))))))............. (-10.10 = -10.50 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:58 2006