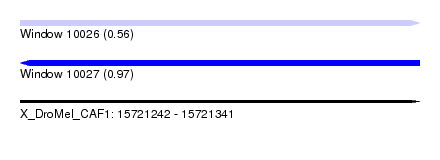
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,721,242 – 15,721,341 |
| Length | 99 |
| Max. P | 0.971218 |

| Location | 15,721,242 – 15,721,341 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15721242 99 + 22224390 ACGGACCAAAUUUUCGAUAGCUGCAGAAAAUCAUUUUCAAGGGGAUAACAUUAUGAUAACUAUCGAAGUAGCGGCCAGUCUCCACUGGUGGCCGCGAUU ...........(((((((((.((((.......(((((....))))).......)).)).)))))))))..(((((((.((......))))))))).... ( -26.04) >DroSec_CAF1 4758 99 + 1 ACGGACCAAAUUUUCGAUAGCUGCAGAAAAUCAUUUUCAAGGGGAUAACAUUAUGAUAACUAUCGAAACAGCGGUCAGUCUCCACUGGUGGCCGCGAUU .(((.(((...(((((((((.((((.......(((((....))))).......)).)).)))))))))(((.((.......)).))).))))))..... ( -23.54) >DroSim_CAF1 4438 99 + 1 ACGGACCAAAUUUUCGAUAGCUGCAGAAAAUCAUUUUCAAGGGGAUAACAUUAUGAUAACUAUCGAAACAGCGGCCAGUCUCCACUGGUGGCCGCGAUU ...........(((((((((.((((.......(((((....))))).......)).)).)))))))))..(((((((.((......))))))))).... ( -26.24) >DroEre_CAF1 4723 99 + 1 ACGGACCAAAUUUUCGAUAACUGCAGAAAAUCAUUUUCAAGGGGAUAACAUUAUGAUAACUAUCGAAAGCGUAGACAGUCUCCACUGGUAUCCGCGAUU .(((((((.((((((..........)))))).........((((((..(.....)....(((((....).))))...))))))..)))..))))..... ( -18.70) >DroYak_CAF1 3960 99 + 1 ACGGACCAAAUUUUCGAUAACUGCAGAAAAUCAUUUUCAAGGGGAUAACAUUAUGAUAACUAUCGAAACAGUGGACUAUCUCCACUGGUGUCCGCGAUU .((((((....((((((((....((.......(((((....))))).......)).....))))))))(((((((.....)))))))).)))))..... ( -25.94) >consensus ACGGACCAAAUUUUCGAUAGCUGCAGAAAAUCAUUUUCAAGGGGAUAACAUUAUGAUAACUAUCGAAACAGCGGACAGUCUCCACUGGUGGCCGCGAUU ...........(((((((((.((((.......(((((....))))).......)).)).)))))))))..(((((((.((......))))))))).... (-20.52 = -20.56 + 0.04)


| Location | 15,721,242 – 15,721,341 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -23.11 |
| Energy contribution | -22.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15721242 99 - 22224390 AAUCGCGGCCACCAGUGGAGACUGGCCGCUACUUCGAUAGUUAUCAUAAUGUUAUCCCCUUGAAAAUGAUUUUCUGCAGCUAUCGAAAAUUUGGUCCGU ....(((((((.....(....))))))))...(((((((((((((((....(((......)))..))))).......))))))))))............ ( -26.61) >DroSec_CAF1 4758 99 - 1 AAUCGCGGCCACCAGUGGAGACUGACCGCUGUUUCGAUAGUUAUCAUAAUGUUAUCCCCUUGAAAAUGAUUUUCUGCAGCUAUCGAAAAUUUGGUCCGU ....(((((((.((((((.......))))))((((((((((((((((....(((......)))..))))).......)))))))))))...))).)))) ( -29.01) >DroSim_CAF1 4438 99 - 1 AAUCGCGGCCACCAGUGGAGACUGGCCGCUGUUUCGAUAGUUAUCAUAAUGUUAUCCCCUUGAAAAUGAUUUUCUGCAGCUAUCGAAAAUUUGGUCCGU ....(((((((.((((((.......))))))((((((((((((((((....(((......)))..))))).......)))))))))))...))).)))) ( -29.21) >DroEre_CAF1 4723 99 - 1 AAUCGCGGAUACCAGUGGAGACUGUCUACGCUUUCGAUAGUUAUCAUAAUGUUAUCCCCUUGAAAAUGAUUUUCUGCAGUUAUCGAAAAUUUGGUCCGU ....(((((..((((((((.....)))))..((((((((((.(((((....(((......)))..))))).....))...))))))))...)))))))) ( -20.60) >DroYak_CAF1 3960 99 - 1 AAUCGCGGACACCAGUGGAGAUAGUCCACUGUUUCGAUAGUUAUCAUAAUGUUAUCCCCUUGAAAAUGAUUUUCUGCAGUUAUCGAAAAUUUGGUCCGU ....((((((..(((((((.....)))))))((((((((((.(((((....(((......)))..))))).....))...)))))))).....)))))) ( -27.50) >consensus AAUCGCGGCCACCAGUGGAGACUGGCCGCUGUUUCGAUAGUUAUCAUAAUGUUAUCCCCUUGAAAAUGAUUUUCUGCAGCUAUCGAAAAUUUGGUCCGU ....((((..((((((((.......))))..((((((((((((((((....(((......)))..))))).......)))))))))))...)))))))) (-23.11 = -22.67 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:55 2006