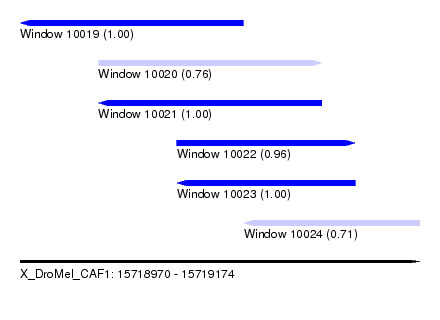
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,718,970 – 15,719,174 |
| Length | 204 |
| Max. P | 0.999263 |

| Location | 15,718,970 – 15,719,084 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -42.42 |
| Consensus MFE | -41.26 |
| Energy contribution | -41.54 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15718970 114 - 22224390 GUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCCAGUAGCAGUACUCAUCUGAGUCUGAAUGCCAAGCAACAGCGCCAGCAGAAACGCCGGAGGGAACGUGCCCAGCGU ((((((..(((.....)))..)))))).((((..(((((.((..(((.((((....)))))))..(((...((....))....))).....)).)))))....))))....... ( -42.60) >DroSec_CAF1 2456 114 - 1 GUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCUAGCAGCAGCACUCAUCUGAGUCUGAAUGCCAAGCAACAGCGCCAGCAGAAACGCCGGAGGGAACGUGCCCAGCGU ((((((..(((.....)))..)))))).((((..((((..((..(((.((((....)))))))..(((...((....))....))).....))..))))....))))....... ( -43.60) >DroSim_CAF1 1810 114 - 1 AUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCCAGCAGCAGCACUCAUCUGAGUCUGAAUGCCAAGCAACAGCGCCAGCAGAAACGCCGGAGGGAACGUGCCCAGCGU .(((((..(((.....)))..)))))..((((..(((((.((..(((.((((....)))))))..(((...((....))....))).....)).)))))....))))....... ( -41.40) >DroEre_CAF1 2426 114 - 1 GUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCCAGCAGCAGCACUCACCUAAGUCUGAAUGCCAAGCAACAGCGCCAGCAGAAACGCCGGAGGGAACGUGCCCAGCGC ((((((..(((.....)))..)))))).((((..(((((.((.((.(((.(((........))).)))...))....((....))......)).)))))....))))....... ( -41.60) >DroYak_CAF1 1785 114 - 1 GUGGCUGCGGGGAACACCAAUGGCCACAGCACCACCUCCAGCAGCAGCACUCACCUGAGUCUGAAUGCCAAGCAACAGCGCCAGCAGAAACGCCGGAGGGAACGUGCCCAGCGU (((((((..((.....))..))))))).((((..(((((.((..(((.((((....)))))))..(((...((....))....))).....)).)))))....))))....... ( -42.90) >consensus GUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCCAGCAGCAGCACUCAUCUGAGUCUGAAUGCCAAGCAACAGCGCCAGCAGAAACGCCGGAGGGAACGUGCCCAGCGU ((((((..(((.....)))..)))))).((((..(((((.((..(((.((((....)))))))..(((...((....))....))).....)).)))))....))))....... (-41.26 = -41.54 + 0.28)

| Location | 15,719,010 – 15,719,124 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 94.21 |
| Mean single sequence MFE | -44.76 |
| Consensus MFE | -39.30 |
| Energy contribution | -40.34 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15719010 114 + 22224390 UGCUUGGCAUUCAGACUCAGAUGAGUACUGCUACUGGAGGUGGUGCUGUGGCCAUUGGUAUUCCCAGCAGCCACCGCUCCUGAUCCCGCUCCGAAGCCGGACUGAUUACCGGCU .((.(((((.....((((....))))..)))))..(((...((.((.(((((..((((.....))))..))))).)).))...))).)).....((((((........)))))) ( -41.90) >DroSec_CAF1 2496 114 + 1 UGCUUGGCAUUCAGACUCAGAUGAGUGCUGCUGCUAGAGGUGGUGCUGUGGCCAUUGGUAUUCCCAGCAGCCACCGCUCCUGAUCCCGCUCCGAAGCCGGACUGACUACCGGCU .((((((((..(((((((....)))).))).)))))).((.((.((.(((((..((((.....))))..))))).)).))....)).)).....((((((........)))))) ( -43.30) >DroSim_CAF1 1850 114 + 1 UGCUUGGCAUUCAGACUCAGAUGAGUGCUGCUGCUGGAGGUGGUGCUGUGGCCAUUGGUAUUCCCAGCAGCCAUUGCUCCUGAUCCCGCUCCGAAGCCGGACUGACUACCGGCU .((..((((((((........)))))))))).((.(((...((.((.(((((..((((.....))))..))))).)).))...))).)).....((((((........)))))) ( -45.50) >DroEre_CAF1 2466 114 + 1 UGCUUGGCAUUCAGACUUAGGUGAGUGCUGCUGCUGGAGGUGGUGCUGUGGCCAUUGGUAUUCCCAGCAGCCACCGCUCCUGAUCCCGCUCCGAAGCCAGUCUGAUUACCGGCA .(((.((...(((((((..(((((((((....)).(((...((.((.(((((..((((.....))))..))))).)).))...))).))))....))))))))))...))))). ( -44.60) >DroYak_CAF1 1825 114 + 1 UGCUUGGCAUUCAGACUCAGGUGAGUGCUGCUGCUGGAGGUGGUGCUGUGGCCAUUGGUGUUCCCCGCAGCCACCGCUCCUGAUCCCGCUCCGAAGCCAGUCUGAAUCCCGGCU .(((.((.(((((((((..((((((((......(.((((.(((((((((((.....((....)))))))).))))))))).)....)))))....)))))))))))))).))). ( -48.50) >consensus UGCUUGGCAUUCAGACUCAGAUGAGUGCUGCUGCUGGAGGUGGUGCUGUGGCCAUUGGUAUUCCCAGCAGCCACCGCUCCUGAUCCCGCUCCGAAGCCGGACUGACUACCGGCU .((..((((((((........)))))))))).((.(((...((.((.(((((..((((.....))))..))))).)).))...))).))......(((((........))))). (-39.30 = -40.34 + 1.04)



| Location | 15,719,010 – 15,719,124 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.21 |
| Mean single sequence MFE | -45.02 |
| Consensus MFE | -42.94 |
| Energy contribution | -42.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15719010 114 - 22224390 AGCCGGUAAUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCGGUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCCAGUAGCAGUACUCAUCUGAGUCUGAAUGCCAAGCA ((((((........)))))).....((.(((...((.((.((((((..(((.....)))..)))))).)).))...))).(((.(((.((((....)))))))..)))...)). ( -45.10) >DroSec_CAF1 2496 114 - 1 AGCCGGUAGUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCGGUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCUAGCAGCAGCACUCAUCUGAGUCUGAAUGCCAAGCA ((((((........))))))..((.((.(((...((.((.((((((..(((.....)))..)))))).)).))...))).))..(((.((((....)))))))....))..... ( -45.10) >DroSim_CAF1 1850 114 - 1 AGCCGGUAGUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCAAUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCCAGCAGCAGCACUCAUCUGAGUCUGAAUGCCAAGCA ((((((........)))))).....((.(((...((.((..(((((..(((.....)))..)))))..)).))...))).(((.(((.((((....)))))))..)))...)). ( -43.50) >DroEre_CAF1 2466 114 - 1 UGCCGGUAAUCAGACUGGCUUCGGAGCGGGAUCAGGAGCGGUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCCAGCAGCAGCACUCACCUAAGUCUGAAUGCCAAGCA .((.((((.(((((((.(((.(...((.(((...((.((.((((((..(((.....)))..)))))).)).))...))).)).).))).........))))))).))))..)). ( -46.10) >DroYak_CAF1 1825 114 - 1 AGCCGGGAUUCAGACUGGCUUCGGAGCGGGAUCAGGAGCGGUGGCUGCGGGGAACACCAAUGGCCACAGCACCACCUCCAGCAGCAGCACUCACCUGAGUCUGAAUGCCAAGCA .((.((.((((((((((((....).((.(((...((.((.(((((((..((.....))..))))))).)).))...))).))...........))..))))))))).))..)). ( -45.30) >consensus AGCCGGUAAUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCGGUGGCUGCUGGGAAUACCAAUGGCCACAGCACCACCUCCAGCAGCAGCACUCAUCUGAGUCUGAAUGCCAAGCA .(((((........)))))......((.(((...((.((.((((((..(((.....)))..)))))).)).))...))).(((.(((.((((....)))))))..)))...)). (-42.94 = -42.98 + 0.04)

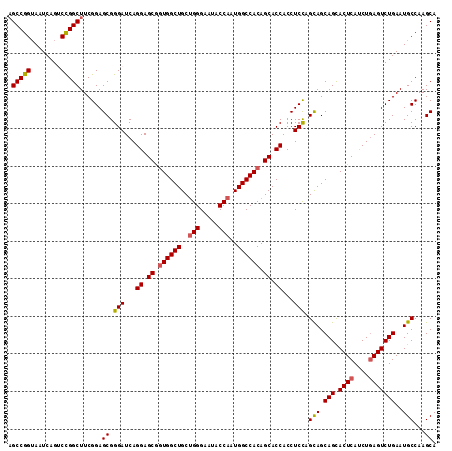
| Location | 15,719,050 – 15,719,141 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -29.72 |
| Energy contribution | -30.17 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.962070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15719050 91 + 22224390 UGGUGCUGUGGCCAUUGGUAUUCCCAGCAGCCACCGCUCCUGAUCCCGCUCCGAAGCCGGACUGAUUACCGGCUAUGCUAGCAGUGCCUCC----------------------- .((..(((((((..((((.....))))..))))).............(((.((.((((((........)))))).))..)))))..))...----------------------- ( -33.10) >DroSim_CAF1 1890 114 + 1 UGGUGCUGUGGCCAUUGGUAUUCCCAGCAGCCAUUGCUCCUGAUCCCGCUCCGAAGCCGGACUGACUACCGGCUAUGCUAGCAGUGCCUCCUCCGCCUCCUACUCCUCCUNNNN .((..(((((((..((((......(((.(((....))).)))........))))((((((........))))))..)))).)))..)).......................... ( -32.44) >DroEre_CAF1 2506 101 + 1 UGGUGCUGUGGCCAUUGGUAUUCCCAGCAGCCACCGCUCCUGAUCCCGCUCCGAAGCCAGUCUGAUUACCGGCAUUGUUAGCAGUGCCUACUCCACCUCCU------------- .((.((.(((((..((((.....))))..))))).)).))(((((..(((........)))..)))))..(((((((....))))))).............------------- ( -32.40) >consensus UGGUGCUGUGGCCAUUGGUAUUCCCAGCAGCCACCGCUCCUGAUCCCGCUCCGAAGCCGGACUGAUUACCGGCUAUGCUAGCAGUGCCUCCUCC_CCUCCU_____________ .((..(((((((..((((.....))))..))))).............(((.((.((((((........)))))).))..)))))..)).......................... (-29.72 = -30.17 + 0.45)

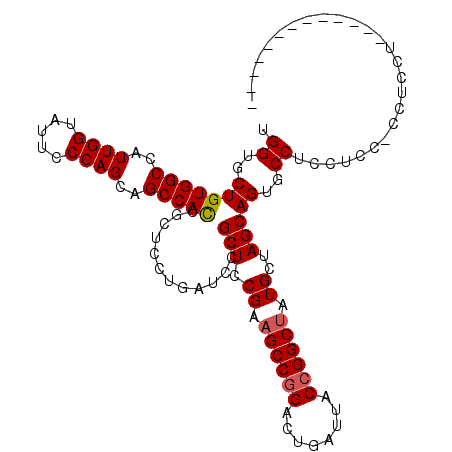

| Location | 15,719,050 – 15,719,141 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -39.39 |
| Energy contribution | -39.83 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15719050 91 - 22224390 -----------------------GGAGGCACUGCUAGCAUAGCCGGUAAUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCGGUGGCUGCUGGGAAUACCAAUGGCCACAGCACCA -----------------------...((..(((((.....((((((........))))))....)))))..)).((.((.((((((..(((.....)))..)))))).)).)). ( -42.40) >DroSim_CAF1 1890 114 - 1 NNNNAGGAGGAGUAGGAGGCGGAGGAGGCACUGCUAGCAUAGCCGGUAGUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCAAUGGCUGCUGGGAAUACCAAUGGCCACAGCACCA ......((...((.((((.((((....(.(((((..((...))..))))))..)))).))))...))....)).((.((..(((((..(((.....)))..)))))..)).)). ( -39.60) >DroEre_CAF1 2506 101 - 1 -------------AGGAGGUGGAGUAGGCACUGCUAACAAUGCCGGUAAUCAGACUGGCUUCGGAGCGGGAUCAGGAGCGGUGGCUGCUGGGAAUACCAAUGGCCACAGCACCA -------------....(((......((..(((((......((((((......)))))).....)))))..))....((.((((((..(((.....)))..)))))).))))). ( -40.50) >consensus _____________AGGAGG_GGAGGAGGCACUGCUAGCAUAGCCGGUAAUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCGGUGGCUGCUGGGAAUACCAAUGGCCACAGCACCA ..........................((..(((((.....((((((........))))))....)))))..)).((.((.((((((..(((.....)))..)))))).)).)). (-39.39 = -39.83 + 0.45)

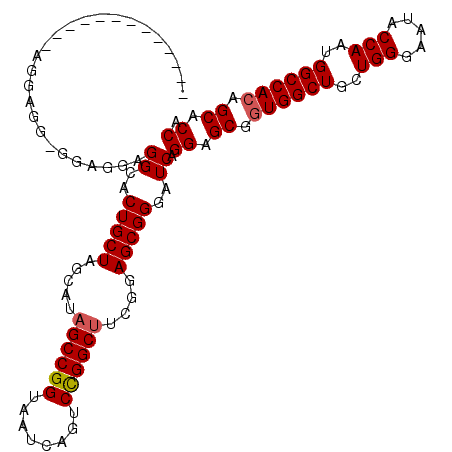

| Location | 15,719,084 – 15,719,174 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 57.60 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -18.69 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15719084 90 - 22224390 UCCACAAUAAUGCCACAGCUGAAUUCCAGUGCG------------------------------GGAGGCACUGCUAGCAUAGCCGGUAAUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCG (((.......((((.(.((((.....))))...------------------------------)..))))(((((.....((((((........))))))....))))).....)))... ( -31.70) >DroSim_CAF1 1924 120 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAGGAGGAGUAGGAGGCGGAGGAGGCACUGCUAGCAUAGCCGGUAGUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCA ..........................................................((....((....(((((.....((((((........))))))....)))))..))....)). ( -22.80) >DroEre_CAF1 2540 102 - 1 UCCACAAUAAUGCCACAGCUGAAUUCCAGUGCGGG------------------AGGAGGUGGAGUAGGCACUGCUAACAAUGCCGGUAAUCAGACUGGCUUCGGAGCGGGAUCAGGAGCG (((.......((((((.(((...((((......))------------------))..)))...)).))))(((((......((((((......)))))).....))))).....)))... ( -32.40) >consensus UCCACAAUAAUGCCACAGCUGAAUUCCAGUGCG____________________AGGAGG_GGAGGAGGCACUGCUAGCAUAGCCGGUAAUCAGUCCGGCUUCGGAGCGGGAUCAGGAGCG ..................................................................((..(((((.....((((((........))))))....)))))..))....... (-18.69 = -18.80 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:52 2006