| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,718,473 – 15,718,632 |
| Length | 159 |
| Max. P | 0.919082 |

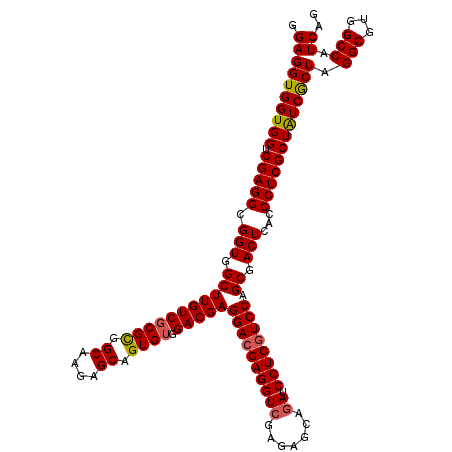
| Location | 15,718,473 – 15,718,592 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -52.96 |
| Consensus MFE | -50.08 |
| Energy contribution | -50.04 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15718473 119 + 22224390 GGAGGUGGUGGUCGAGGCGGUGGCUUGUCGCGCGGCAAGAGCAGUGUGGACGAGGACGAGGUCGAGAGCAGAUCCUCGUCCAGCGACUCACCCUCGCUAUCGCUAGGCGUGGCCAUCAA .((((((((((.(((((.(((.(((((((((((.((....)).)))).)))))((((((((((.......)).)))))))).)).)))...))))))))))))).(((...))).)).. ( -55.10) >DroSec_CAF1 1965 119 + 1 GGAGGUGGUGGUCGAGGCGGUGGCUUGUCGCGCGGCAAGAGCAGUGUGGACGAGGACGAGGUCGAGAGCAGAUCCUCGUCCAGCGACUCACCCUCGCUAUCGCUAGGCGUGGCCAUCAG .((((((((((.(((((.(((.(((((((((((.((....)).)))).)))))((((((((((.......)).)))))))).)).)))...))))))))))))).(((...))).)).. ( -55.10) >DroSim_CAF1 1313 119 + 1 GGAGGUGGUGGUCGAGGCGGUGGCUUGUCGCGCGGCAAGAGCAGUGUGGACGAGGACGAGGUCGAGAGCAGAUCCUCGUCCAGCGACUCACCCUCGCUAUCGCUAGGCGUGGCCAUCAG .((((((((((.(((((.(((.(((((((((((.((....)).)))).)))))((((((((((.......)).)))))))).)).)))...))))))))))))).(((...))).)).. ( -55.10) >DroEre_CAF1 1935 119 + 1 GGAGGUGGUGGUCGAGGCGGUGGCUUGUCGCGUGGCAAGAGCAGUGUGGACGAGGACGAGGUAGAGAGCAGAUCCUCGUCCAGCGACUCACCCUCGCUGUCACUAGGCGUGGCCAUCAG .....(((((((((.(((((.((...(((((...((....))....(((((((((((...((.....)).).)))))))))))))))...)).)))))(((....))).))))))))). ( -51.70) >DroYak_CAF1 1291 119 + 1 GGAGGUGGUGGUCGAGGCGGUGGCUUGUCCCGUGGCAAGAGCAGUGUCGACGAGGACGAGGUCGAUAGCAGAUCCUCAUCCAGCGACUCACCCUCGCUAUCACUAGGCGUGGCCAUCAG .((((((((((.(((((.(((.(((.(((((((((((.......)))).))).))))((((((.......)).))))....))).)))...))))))))))))).(((...))).)).. ( -47.80) >consensus GGAGGUGGUGGUCGAGGCGGUGGCUUGUCGCGCGGCAAGAGCAGUGUGGACGAGGACGAGGUCGAGAGCAGAUCCUCGUCCAGCGACUCACCCUCGCUAUCGCUAGGCGUGGCCAUCAG .((((((((((.(((((.(((.(((((((((((.((....)).)))).)))))((((((((((.......)).)))))))).)).)))...))))))))))))).(((...))).)).. (-50.08 = -50.04 + -0.04)


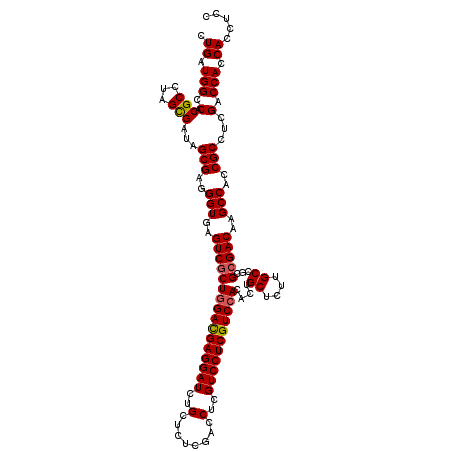
| Location | 15,718,473 – 15,718,592 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -43.24 |
| Consensus MFE | -40.36 |
| Energy contribution | -40.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15718473 119 - 22224390 UUGAUGGCCACGCCUAGCGAUAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCGACCUCGUCCUCGUCCACACUGCUCUUGCCGCGCGACAAGCCACCGCCUCGACCACCACCUCC ((((.((((((.(((.((....)).)))))).((((((((((((((((..(........)..)))))))))))....((....))...)))))........)))))))........... ( -43.90) >DroSec_CAF1 1965 119 - 1 CUGAUGGCCACGCCUAGCGAUAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCGACCUCGUCCUCGUCCACACUGCUCUUGCCGCGCGACAAGCCACCGCCUCGACCACCACCUCC .((.(((.(.(((...)))...(((..(((..((((((((((((((((..(........)..)))))))))))....((....))...)))))..)))..)))...).))).))..... ( -43.70) >DroSim_CAF1 1313 119 - 1 CUGAUGGCCACGCCUAGCGAUAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCGACCUCGUCCUCGUCCACACUGCUCUUGCCGCGCGACAAGCCACCGCCUCGACCACCACCUCC .((.(((.(.(((...)))...(((..(((..((((((((((((((((..(........)..)))))))))))....((....))...)))))..)))..)))...).))).))..... ( -43.70) >DroEre_CAF1 1935 119 - 1 CUGAUGGCCACGCCUAGUGACAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCUACCUCGUCCUCGUCCACACUGCUCUUGCCACGCGACAAGCCACCGCCUCGACCACCACCUCC .((.(((.(((.....)))...(((..(((..((((((((((((((((..(........)..)))))))))))....((....))...)))))..)))..))).....))).))..... ( -43.60) >DroYak_CAF1 1291 119 - 1 CUGAUGGCCACGCCUAGUGAUAGCGAGGGUGAGUCGCUGGAUGAGGAUCUGCUAUCGACCUCGUCCUCGUCGACACUGCUCUUGCCACGGGACAAGCCACCGCCUCGACCACCACCUCC ..(.((((....(((.(((...(((((((((.(((((.((((((((.((.......))))))))))..).)))).).))))))))))))))....)))).).................. ( -41.30) >consensus CUGAUGGCCACGCCUAGCGAUAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCGACCUCGUCCUCGUCCACACUGCUCUUGCCGCGCGACAAGCCACCGCCUCGACCACCACCUCC .((.(((.(.(((...)))...(((..(((..((((((((((((((((..(........)..)))))))))))....((....))...)))))..)))..)))...).))).))..... (-40.36 = -40.36 + -0.00)


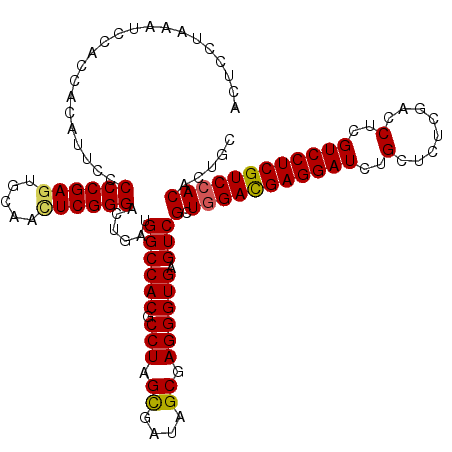
| Location | 15,718,513 – 15,718,632 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -41.02 |
| Consensus MFE | -38.94 |
| Energy contribution | -38.82 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15718513 119 - 22224390 ACUCCUAAAUCCACCACAUUCCCCCGAGUGCAACUCGGGAUUGAUGGCCACGCCUAGCGAUAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCGACCUCGUCCUCGUCCACACUGC ......................((((((.....))))))......((((((.(((.((....)).)))))).)))(.(((((((((((..(........)..))))))))))))..... ( -42.80) >DroSec_CAF1 2005 119 - 1 ACUCCUAAAUCCACCACAUUCCCCCGAGUGCAACUCGGGUCUGAUGGCCACGCCUAGCGAUAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCGACCUCGUCCUCGUCCACACUGC ......................((((((.....))))))......((((((.(((.((....)).)))))).)))(.(((((((((((..(........)..))))))))))))..... ( -42.30) >DroSim_CAF1 1353 119 - 1 ACUCCUAAAUCCACCACAUUCCCCCGAGUGCAACUCGGGUCUGAUGGCCACGCCUAGCGAUAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCGACCUCGUCCUCGUCCACACUGC ......................((((((.....))))))......((((((.(((.((....)).)))))).)))(.(((((((((((..(........)..))))))))))))..... ( -42.30) >DroEre_CAF1 1975 119 - 1 ACUCCUAAAUCCACCACAUUCCCCCGAAUGCAACUCGGGACUGAUGGCCACGCCUAGUGACAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCUACCUCGUCCUCGUCCACACUGC ...........((((.((((..(((((.......)))))...))))(((((.....)))...))...))))(((.(.(((((((((((..(........)..))))))))))))))).. ( -37.70) >DroYak_CAF1 1331 119 - 1 ACUCCUAAAUCCACCACAUUCCCCCGAGUGCAAUUCGGGACUGAUGGCCACGCCUAGUGAUAGCGAGGGUGAGUCGCUGGAUGAGGAUCUGCUAUCGACCUCGUCCUCGUCGACACUGC ...........((((.((((..(((((((...)))))))...))))....(((.((....)))))..)))).(((((.((((((((.((.......))))))))))..).))))..... ( -40.00) >consensus ACUCCUAAAUCCACCACAUUCCCCCGAGUGCAACUCGGGACUGAUGGCCACGCCUAGCGAUAGCGAGGGUGAGUCGCUGGACGAGGAUCUGCUCUCGACCUCGUCCUCGUCCACACUGC ......................((((((.....))))))......((((((.(((.((....)).)))))).)))(.(((((((((((..(........)..))))))))))))..... (-38.94 = -38.82 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:46 2006