
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,718,155 – 15,718,313 |
| Length | 158 |
| Max. P | 0.999378 |

| Location | 15,718,155 – 15,718,274 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.90 |
| Mean single sequence MFE | -36.59 |
| Consensus MFE | -35.32 |
| Energy contribution | -35.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
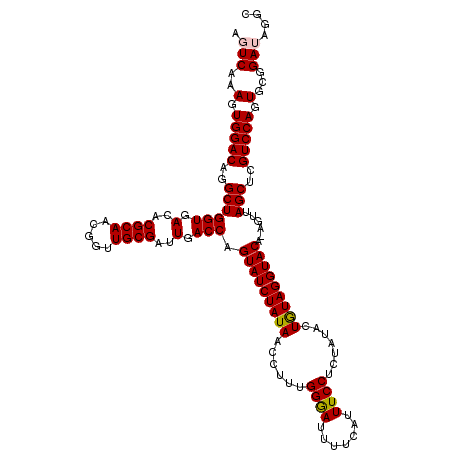
>X_DroMel_CAF1 15718155 119 + 22224390 AGUCAAAGUGGACAGGCUGGUGACACGCAACGGUUGCGAUUGACCAGUAUCUAUAACCUUUGGGAUUUUCAUUUCCUCUAUACUGUAGGUACA-AGUUAGCUCGUCCAGUGCGGAUAGAC .(((..(.(((((..((((((.(..((((.....))))..).))).(((((((((......((((........))))......))))))))).-....)))..))))).)...))).... ( -37.40) >DroSec_CAF1 1647 118 + 1 AGUCAAAGUGGACAGGCUGGUGACACGCAACGGCUGCGAUUGACCAGUAUCUAUAACCUUUGGAAUUU-CA-UUCCUCUAUGCUGUAGGUACAAAGUUAGCUCGUCCAGUGCGGAUAGGC .(((..(.(((((..((((((.(..((((.....))))..).))).(((((((((......(((((..-.)-)))).......)))))))))......)))..))))).)...))).... ( -36.62) >DroSim_CAF1 995 119 + 1 AGUCAAAGUGGACAGGCUGGUGACACGCAACGGCUGCGAUUGACCAGUAUCUAUAACCUUUGGAAUUUUCAUUUCCUCUAUACUGUAGGUACA-AGUUAGCUCGUCCAGUGCGGAUAGGC .(((..(.(((((..((((((.(..((((.....))))..).))).(((((((((......((((.......)))).......))))))))).-....)))..))))).)...))).... ( -36.92) >DroEre_CAF1 1626 119 + 1 AGUCAAAGUGGACAGGCUGGUGACACGCAACGGUUGCGAUUGACCAGUAUCUAUAACCUUUGGGAUUUUCAUUUCCUCUAUACUAUAGGUACA-AGUUAGCUCGUCCAGUGCGGAAAGGC ..((..(.(((((..((((((.(..((((.....))))..).))).(((((((((......((((........))))......))))))))).-....)))..))))).)...))..... ( -36.00) >DroYak_CAF1 982 119 + 1 GGUCAAAGUGGACAGGCUGGUGACACGCAACGGUUGCGAUUGACCAGUAUCUAUAACCUUUGGGAUUUUCAUUUCCUCUAUACUGUAGGUACA-AGUUAGCUCGUCCAGUGCGGAAAGUC ..((..(.(((((..((((((.(..((((.....))))..).))).(((((((((......((((........))))......))))))))).-....)))..))))).)...))..... ( -36.00) >consensus AGUCAAAGUGGACAGGCUGGUGACACGCAACGGUUGCGAUUGACCAGUAUCUAUAACCUUUGGGAUUUUCAUUUCCUCUAUACUGUAGGUACA_AGUUAGCUCGUCCAGUGCGGAUAGGC .(((..(.(((((..((((((.(..((((.....))))..).))).(((((((((......((((.......)))).......)))))))))......)))..))))).)...))).... (-35.32 = -35.32 + 0.00)

| Location | 15,718,195 – 15,718,313 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.37 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -28.54 |
| Energy contribution | -28.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.999378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
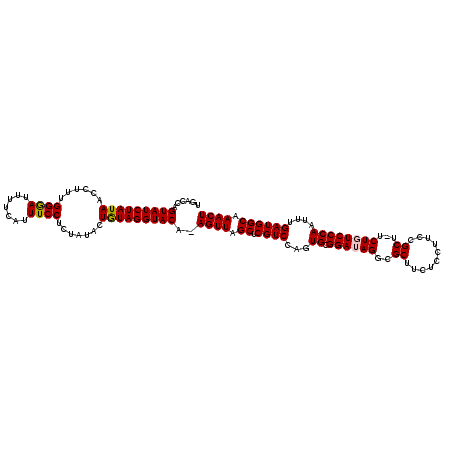
>X_DroMel_CAF1 15718195 118 + 22224390 UGACCAGUAUCUAUAACCUUUGGGAUUUUCAUUUCCUCUAUACUGUAGGUACA-AGUUAGCUCGUCCAGUGCGGAUAGACGCUUCUCCUUCCGCU-UCUGUCCCAAUUUGAUGGCAAACU ......(((((((((......((((........))))......))))))))).-((((.((.((((...((.(((((((.((..........)).-)))))))))....)))))).)))) ( -34.40) >DroSec_CAF1 1687 118 + 1 UGACCAGUAUCUAUAACCUUUGGAAUUU-CA-UUCCUCUAUGCUGUAGGUACAAAGUUAGCUCGUCCAGUGCGGAUAGGCGCUUCUCCUUCCGCUUUCUGUCCCAAUUUGAUGGCAAACU ......(((((((((......(((((..-.)-)))).......)))))))))..((((.((.(((((((.(((((.(((.......))))))))...))).........)))))).)))) ( -31.02) >DroSim_CAF1 1035 118 + 1 UGACCAGUAUCUAUAACCUUUGGAAUUUUCAUUUCCUCUAUACUGUAGGUACA-AGUUAGCUCGUCCAGUGCGGAUAGGCGCUUCUCCUUCCGCU-UCUGUCCCAAUUUGAUGGCAAACU ......(((((((((......((((.......)))).......))))))))).-((((.((.((((...((.(((((((.((..........)).-)))))))))....)))))).)))) ( -32.82) >DroEre_CAF1 1666 118 + 1 UGACCAGUAUCUAUAACCUUUGGGAUUUUCAUUUCCUCUAUACUAUAGGUACA-AGUUAGCUCGUCCAGUGCGGAAAGGCGCUUUUCUUUUCGCU-UCUGUCCCAAUUUGAUGGCAAACU ......(((((((((......((((........))))......))))))))).-((((.((.(((((((.(((((((((......))))))))).-.))).........)))))).)))) ( -32.30) >DroYak_CAF1 1022 118 + 1 UGACCAGUAUCUAUAACCUUUGGGAUUUUCAUUUCCUCUAUACUGUAGGUACA-AGUUAGCUCGUCCAGUGCGGAAAGUCGCUUCUCCUUCCGCU-UCUGUCCCAAUUUGAUGGCAAACU ......(((((((((......((((........))))......))))))))).-((((.((.(((((((.((((((.(........).)))))).-.))).........)))))).)))) ( -30.70) >consensus UGACCAGUAUCUAUAACCUUUGGGAUUUUCAUUUCCUCUAUACUGUAGGUACA_AGUUAGCUCGUCCAGUGCGGAUAGGCGCUUCUCCUUCCGCU_UCUGUCCCAAUUUGAUGGCAAACU ......(((((((((......((((.......)))).......)))))))))..((((.((.((((...((.((((((..((..........))...))))))))....)))))).)))) (-28.54 = -28.54 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:43 2006