
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,717,887 – 15,718,035 |
| Length | 148 |
| Max. P | 0.999840 |

| Location | 15,717,887 – 15,717,995 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -42.16 |
| Consensus MFE | -40.50 |
| Energy contribution | -40.54 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.22 |
| SVM RNA-class probability | 0.999840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15717887 108 + 22224390 GGUCUUGCUGCUGCCCAUAUUCGGAUCGCCAAUGAUCCCUGGGCUCGGCUUCCGUCUCCAGAACAUUGCCCACCAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGG (((((..((((((((.......((((((....))))))(((((..(((...)))...)).((((((........)))))).)))..))))))))..)))))....... ( -42.00) >DroSec_CAF1 1379 108 + 1 GGUCUUGCUGCUGCCCAUAUUCGGAUCGCCAGUGAUCCCUGGGCUCGGCUUCCGUCUCCAGAACAUUGCCGACCAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGG (((((..((((((((.......((((((....))))))...((.(((((.....((....)).....)))))))............))))))))..)))))....... ( -44.10) >DroSim_CAF1 727 108 + 1 GGUCUUGCUGCUGCCCAUAUUCGGAUCGCCAGUGAUCCCUGGGCUCGGCUUCCGUCUCCAGAACAUUGCCGACCAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGG (((((..((((((((.......((((((....))))))...((.(((((.....((....)).....)))))))............))))))))..)))))....... ( -44.10) >DroEre_CAF1 1370 105 + 1 GGUCUUGCUGCUGCCCAUAUUCGGAUCGCCAAUGAUCCCUGGGCUCGGUUUCCGUCUCCAGAACAUUGCC---CAUGUUCACAGAUGGGAGCAGGUAGACCACGUUGG (((((..((((((((((.....((((((....)))))).)))))......(((((((...((((((....---.))))))..))))))))))))..)))))....... ( -42.00) >DroYak_CAF1 726 105 + 1 GGUCUUGCUGCUGCCCAUACUCGGAUCGCCAAUGAUCUCUGGGCUCGGUUUCCGUCUCCAGAACACUGCC---CAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGG (((((..((((((((........(((((....)))))((((((..(((...)))...)).(((((.....---..))))).)))).))))))))..)))))....... ( -38.60) >consensus GGUCUUGCUGCUGCCCAUAUUCGGAUCGCCAAUGAUCCCUGGGCUCGGCUUCCGUCUCCAGAACAUUGCC_ACCAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGG (((((..((((((((((.....((((((....)))))).))))).......((((((...((((((........))))))..)))))).)))))..)))))....... (-40.50 = -40.54 + 0.04)



| Location | 15,717,887 – 15,717,995 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -45.72 |
| Consensus MFE | -38.56 |
| Energy contribution | -38.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15717887 108 - 22224390 CCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUGGUGGGCAAUGUUCUGGAGACGGAAGCCGAGCCCAGGGAUCAUUGGCGAUCCGAAUAUGGGCAGCAGCAAGACC .......(((((...((((((((((((....)).))))(((....(((((....))))))))..(((((.(((((......))))).....))))).))))))))))) ( -46.30) >DroSec_CAF1 1379 108 - 1 CCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUGGUCGGCAAUGUUCUGGAGACGGAAGCCGAGCCCAGGGAUCACUGGCGAUCCGAAUAUGGGCAGCAGCAAGACC .......(((((..((((((((..(((.......(((((((....(((((....)))))))))).)))))(((((......))))).......))))))))..))))) ( -46.81) >DroSim_CAF1 727 108 - 1 CCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUGGUCGGCAAUGUUCUGGAGACGGAAGCCGAGCCCAGGGAUCACUGGCGAUCCGAAUAUGGGCAGCAGCAAGACC .......(((((..((((((((..(((.......(((((((....(((((....)))))))))).)))))(((((......))))).......))))))))..))))) ( -46.81) >DroEre_CAF1 1370 105 - 1 CCAACGUGGUCUACCUGCUCCCAUCUGUGAACAUG---GGCAAUGUUCUGGAGACGGAAACCGAGCCCAGGGAUCAUUGGCGAUCCGAAUAUGGGCAGCAGCAAGACC .......(((((..((((((((((.........((---(((....(((((....))))).....))))).(((((......)))))....))))).)))))..))))) ( -44.60) >DroYak_CAF1 726 105 - 1 CCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUG---GGCAGUGUUCUGGAGACGGAAACCGAGCCCAGAGAUCAUUGGCGAUCCGAGUAUGGGCAGCAGCAAGACC .......(((((..((((((((...........((---(((.(..(((((....)))))..)..)))))(.((((......))))).......))))))))..))))) ( -44.10) >consensus CCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUGGU_GGCAAUGUUCUGGAGACGGAAGCCGAGCCCAGGGAUCAUUGGCGAUCCGAAUAUGGGCAGCAGCAAGACC .......(((((..((((((((................(((....(((((....))))).....)))...(((((......))))).......))))))))..))))) (-38.56 = -38.96 + 0.40)



| Location | 15,717,915 – 15,718,035 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -44.92 |
| Consensus MFE | -40.98 |
| Energy contribution | -40.58 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
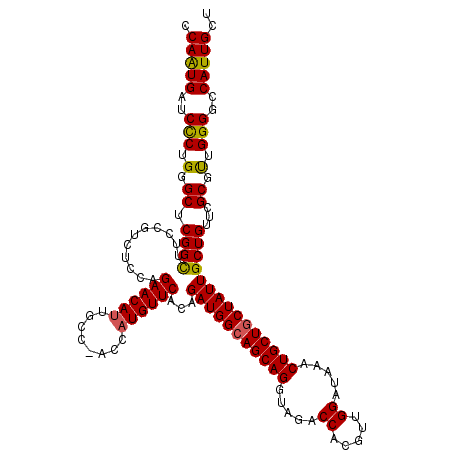
>X_DroMel_CAF1 15717915 120 + 22224390 CCAAUGAUCCCUGGGCUCGGCUUCCGUCUCCAGAACAUUGCCCACCAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGGAUAAACUGCUGCUAUUGCUGUUCGCGUUGGGGCCAUUGCU .(((((....(((((..(((...)))..)))))......((((.(.((((..((((((((((((((.....((.....)).....))))))))))..))))..)))).)))))))))).. ( -45.60) >DroSec_CAF1 1407 120 + 1 CCAGUGAUCCCUGGGCUCGGCUUCCGUCUCCAGAACAUUGCCGACCAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGGAUAAACUGCUGCUAUUGCUGUUCGCGUUGGGGCCAUUGCU .(((((..(((..((.(((((.....((....)).....)))))))((((..((((((((((((((.....((.....)).....))))))))))..))))..)))).)))..))))).. ( -46.70) >DroSim_CAF1 755 120 + 1 CCAGUGAUCCCUGGGCUCGGCUUCCGUCUCCAGAACAUUGCCGACCAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGGAUAAACUGCUGCUAUUGCUGUUCGCGUUGGGGCCAUUGCU .(((((..(((..((.(((((.....((....)).....)))))))((((..((((((((((((((.....((.....)).....))))))))))..))))..)))).)))..))))).. ( -46.70) >DroEre_CAF1 1398 117 + 1 CCAAUGAUCCCUGGGCUCGGUUUCCGUCUCCAGAACAUUGCC---CAUGUUCACAGAUGGGAGCAGGUAGACCACGUUGGAUAAACUGCUGCUAUUGCUGUUCGCGCUGGGACCAUUGCU .(((((.((((.(.((.((((.(((((((...((((((....---.))))))..)))))))(((((.....((.....)).....)))))......))))...)).).)))).))))).. ( -39.90) >DroYak_CAF1 754 117 + 1 CCAAUGAUCUCUGGGCUCGGUUUCCGUCUCCAGAACACUGCC---CAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGGAUAAACUGCUGCUAUUGCUGUUCGCAUUGGGGCCAUUGCU .(((((...((((((..(((...)))..)))))).....(((---(((((..((((((((((((((.....((.....)).....))))))))))..))))..))))..))))))))).. ( -45.70) >consensus CCAAUGAUCCCUGGGCUCGGCUUCCGUCUCCAGAACAUUGCC_ACCAUGUUCACAGAUGGCAGCAGGUAGACCACGUUGGAUAAACUGCUGCUAUUGCUGUUCGCGUUGGGGCCAUUGCU .(((((..(((.(.((.((((...........((((((........))))))...(((((((((((.....((.....)).....)))))))))))))))...)).).)))..))))).. (-40.98 = -40.58 + -0.40)

| Location | 15,717,915 – 15,718,035 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -43.26 |
| Consensus MFE | -36.02 |
| Energy contribution | -36.82 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15717915 120 - 22224390 AGCAAUGGCCCCAACGCGAACAGCAAUAGCAGCAGUUUAUCCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUGGUGGGCAAUGUUCUGGAGACGGAAGCCGAGCCCAGGGAUCAUUGG ..(((((..(((.......((((..((.(((((((.....((.....)).....))))))).))))))........(((((...((((((....))).)))...))))))))..))))). ( -45.30) >DroSec_CAF1 1407 120 - 1 AGCAAUGGCCCCAACGCGAACAGCAAUAGCAGCAGUUUAUCCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUGGUCGGCAAUGUUCUGGAGACGGAAGCCGAGCCCAGGGAUCACUGG ..((.((..(((.......((((..((.(((((((.....((.....)).....))))))).)))))).....((((((((....(((((....))))))))))..))))))..)).)). ( -42.70) >DroSim_CAF1 755 120 - 1 AGCAAUGGCCCCAACGCGAACAGCAAUAGCAGCAGUUUAUCCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUGGUCGGCAAUGUUCUGGAGACGGAAGCCGAGCCCAGGGAUCACUGG ..((.((..(((.......((((..((.(((((((.....((.....)).....))))))).)))))).....((((((((....(((((....))))))))))..))))))..)).)). ( -42.70) >DroEre_CAF1 1398 117 - 1 AGCAAUGGUCCCAGCGCGAACAGCAAUAGCAGCAGUUUAUCCAACGUGGUCUACCUGCUCCCAUCUGUGAACAUG---GGCAAUGUUCUGGAGACGGAAACCGAGCCCAGGGAUCAUUGG ..((((((((((..((((....((....))(((((.....((.....)).....)))))......))))....((---(((....(((((....))))).....))))))))))))))). ( -44.60) >DroYak_CAF1 754 117 - 1 AGCAAUGGCCCCAAUGCGAACAGCAAUAGCAGCAGUUUAUCCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUG---GGCAGUGUUCUGGAGACGGAAACCGAGCCCAGAGAUCAUUGG ..(((((..(.(.(((...((((..((.(((((((.....((.....)).....))))))).))))))...)))(---(((.(..(((((....)))))..)..)))).).)..))))). ( -41.00) >consensus AGCAAUGGCCCCAACGCGAACAGCAAUAGCAGCAGUUUAUCCAACGUGGUCUACCUGCUGCCAUCUGUGAACAUGGU_GGCAAUGUUCUGGAGACGGAAGCCGAGCCCAGGGAUCAUUGG ..(((((..(((.......((((..((.(((((((.....((.....)).....))))))).))))))..........(((....(((((....))))).....)))..)))..))))). (-36.02 = -36.82 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:41 2006