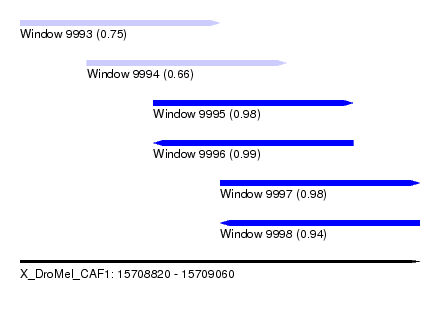
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,708,820 – 15,709,060 |
| Length | 240 |
| Max. P | 0.991413 |

| Location | 15,708,820 – 15,708,940 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -53.23 |
| Consensus MFE | -51.59 |
| Energy contribution | -50.93 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15708820 120 + 22224390 CGCCCUUAUCACCGAGGGCGACUUUCGAUUCCCGCAUGGCAAUGGCCUGGGAUUGGACAUGGGCGCCGAGGGCGGUGCCUGCGGCCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAU (((((((......)))))))..((((...(((((...(((....))))))))..(((((.((((((((....))))))))..((((.......))))..........))))))))).... ( -52.40) >DroSec_CAF1 20006 120 + 1 CGCCCUCAUCACCGAGGGCGACUUUCGGUUCCCGCAUGGCAAUGGCCUGGGAUUGGACAUGGGCGCCGAGGGCGGUGCCUGCGGCCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAU (((((((......)))))))..((((((....((.(((((..(((((((.....((.(..((((((((....))))))))..).))....)).)))))...))))).)).)))))).... ( -55.80) >DroEre_CAF1 19906 120 + 1 AGCCCUCAUCACCGAGGGCGACUUUCGAUUCCCGCAUGGUAAUGGCCUGGGAUUGGACAUGGGCGCCGAGGGCGGUGCCUGCGGUCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAU .((((((......))))))((((((((((.((((...(((....)))))))))))))...((((((((....))))))))..))))((((((.(((.....)))...))))))....... ( -51.50) >consensus CGCCCUCAUCACCGAGGGCGACUUUCGAUUCCCGCAUGGCAAUGGCCUGGGAUUGGACAUGGGCGCCGAGGGCGGUGCCUGCGGCCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAU .((((((......))))))...((((...(((((...(((....))))))))..(((((.((((((((....))))))))..((((.......))))..........))))))))).... (-51.59 = -50.93 + -0.66)

| Location | 15,708,860 – 15,708,980 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -57.37 |
| Consensus MFE | -56.87 |
| Energy contribution | -56.87 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15708860 120 + 22224390 AAUGGCCUGGGAUUGGACAUGGGCGCCGAGGGCGGUGCCUGCGGCCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAUGCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUC ..(((((..(((........((((((((....))))))))((((...(....)(((((...(((((..(((((.........))))))))))..))))).)))).)).)..))))).... ( -56.80) >DroSec_CAF1 20046 120 + 1 AAUGGCCUGGGAUUGGACAUGGGCGCCGAGGGCGGUGCCUGCGGCCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAUGCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUC ..(((((..(((........((((((((....))))))))((((...(....)(((((...(((((..(((((.........))))))))))..))))).)))).)).)..))))).... ( -56.80) >DroEre_CAF1 19946 120 + 1 AAUGGCCUGGGAUUGGACAUGGGCGCCGAGGGCGGUGCCUGCGGUCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAUGCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUU ..(((((..((((..(((..((((((((....))))))))...)))..)....(((((...(((((..(((((.........))))))))))..)))))......)).)..))))).... ( -58.50) >consensus AAUGGCCUGGGAUUGGACAUGGGCGCCGAGGGCGGUGCCUGCGGCCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAUGCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUC ..(((((..(((........((((((((....))))))))((((...(....)(((((...(((((..(((((.........))))))))))..))))).)))).)).)..))))).... (-56.87 = -56.87 + 0.00)


| Location | 15,708,900 – 15,709,020 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -52.53 |
| Consensus MFE | -52.75 |
| Energy contribution | -52.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.58 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15708900 120 + 22224390 GCGGCCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAUGCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUCCACUUGACAAAUAAUGGAGCUGGAUGCGCCGGCAACAUUG ......((((((.(((.....)))...)))))).((((.((((((.((((((((.(((....))))))))...(((((((((.(........).)))))))))..))))))))).)))). ( -53.50) >DroSec_CAF1 20086 120 + 1 GCGGCCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAUGCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUCCACUUGACAAAUAAUGGAGCUGGAUGCGCCGGCAACAUUG ......((((((.(((.....)))...)))))).((((.((((((.((((((((.(((....))))))))...(((((((((.(........).)))))))))..))))))))).)))). ( -53.50) >DroEre_CAF1 19986 120 + 1 GCGGUCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAUGCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUUCACUUGACAAAUAAUGGAGCUGGAUGCGCCGGCAACAUUG ......((((((.(((.....)))...)))))).((((.((((((.((((((((.(((....))))))))...(((((((((.(........).)))))))))..))))))))).)))). ( -50.60) >consensus GCGGCCUGGACACGGCCAAUAGCCAUCUGUCCGAAAUGAUGCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUCCACUUGACAAAUAAUGGAGCUGGAUGCGCCGGCAACAUUG ......((((((.(((.....)))...)))))).((((.((((((.((((((((.(((....))))))))...(((((((((.(........).)))))))))..))))))))).)))). (-52.75 = -52.53 + -0.22)



| Location | 15,708,900 – 15,709,020 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -54.93 |
| Consensus MFE | -54.97 |
| Energy contribution | -55.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15708900 120 - 22224390 CAAUGUUGCCGGCGCAUCCAGCUCCAUUAUUUGUCAAGUGGAGCUGGCCCAGCGAGGCGGUGGCCAUCGCCACAUCCGGCAUCAUUUCGGACAGAUGGCUAUUGGCCGUGUCCAGGCCGC .((((.(((((((((..(((((((((((........)))))))))))....))).((((((....))))))....)))))).))))..(((((..((((.....)))))))))....... ( -57.20) >DroSec_CAF1 20086 120 - 1 CAAUGUUGCCGGCGCAUCCAGCUCCAUUAUUUGUCAAGUGGAGCUGGCCCAGCGAGGCGGUGGCCAUCGCCACAUCCGGCAUCAUUUCGGACAGAUGGCUAUUGGCCGUGUCCAGGCCGC .((((.(((((((((..(((((((((((........)))))))))))....))).((((((....))))))....)))))).))))..(((((..((((.....)))))))))....... ( -57.20) >DroEre_CAF1 19986 120 - 1 CAAUGUUGCCGGCGCAUCCAGCUCCAUUAUUUGUCAAGUGAAGCUGGCCCAGCGAGGCGGUGGCCAUCGCCACAUCCGGCAUCAUUUCGGACAGAUGGCUAUUGGCCGUGUCCAGACCGC .((((.(((((((((..((((((.((((........)))).))))))....))).((((((....))))))....)))))).))))..(((((..((((.....)))))))))....... ( -50.40) >consensus CAAUGUUGCCGGCGCAUCCAGCUCCAUUAUUUGUCAAGUGGAGCUGGCCCAGCGAGGCGGUGGCCAUCGCCACAUCCGGCAUCAUUUCGGACAGAUGGCUAUUGGCCGUGUCCAGGCCGC .((((.(((((((((..(((((((((((........)))))))))))....))).((((((....))))))....)))))).))))..(((((..((((.....)))))))))....... (-54.97 = -55.30 + 0.33)


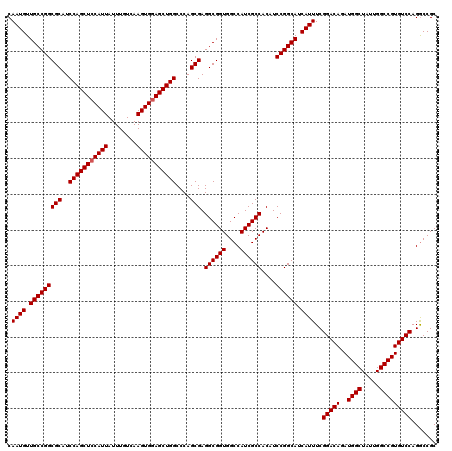
| Location | 15,708,940 – 15,709,060 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -54.47 |
| Consensus MFE | -52.81 |
| Energy contribution | -52.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15708940 120 + 22224390 GCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUCCACUUGACAAAUAAUGGAGCUGGAUGCGCCGGCAACAUUGGCGGGCGCGCAGUGAGCAGCAAUAACGACAUGUCCGAGAG ..((((((((((....))))).(((((((((..(((((((((.(........).)))))))))..(((((.((.......)).))))).)))))))..))...........))))).... ( -56.70) >DroSec_CAF1 20126 120 + 1 GCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUCCACUUGACAAAUAAUGGAGCUGGAUGCGCCGGCAACAUUGGCGGGCGCGCAGUGAGCAGCAAUAACGACAUGUCCGAGAG ..((((((((((....))))).(((((((((..(((((((((.(........).)))))))))..(((((.((.......)).))))).)))))))..))...........))))).... ( -56.70) >DroEre_CAF1 20026 120 + 1 GCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUUCACUUGACAAAUAAUGGAGCUGGAUGCGCCGGCAACAUUGGCGGACGCGCAGUGAGCAGUAAUAACGACAUGUCCGAGAG ..((((((((((....))))).((.((((((..(((((((((.(........).)))))))))..((((((.((....)).))).))).))))))))..............))))).... ( -50.00) >consensus GCCGGAUGUGGCGAUGGCCACCGCCUCGCUGGGCCAGCUCCACUUGACAAAUAAUGGAGCUGGAUGCGCCGGCAACAUUGGCGGGCGCGCAGUGAGCAGCAAUAACGACAUGUCCGAGAG ..((((((((((....))))).(((((((((..(((((((((.(........).)))))))))..(((((.((.......)).))))).)))))))..))...........))))).... (-52.81 = -52.70 + -0.11)

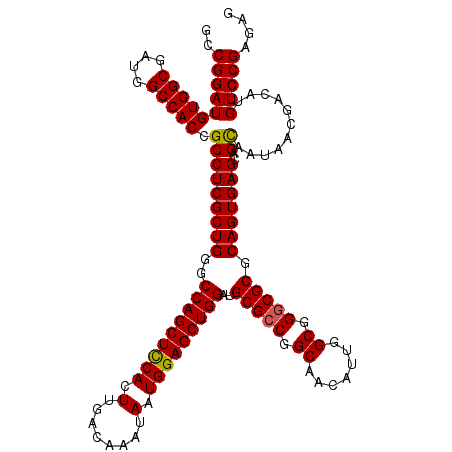

| Location | 15,708,940 – 15,709,060 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -50.00 |
| Consensus MFE | -49.15 |
| Energy contribution | -49.60 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15708940 120 - 22224390 CUCUCGGACAUGUCGUUAUUGCUGCUCACUGCGCGCCCGCCAAUGUUGCCGGCGCAUCCAGCUCCAUUAUUUGUCAAGUGGAGCUGGCCCAGCGAGGCGGUGGCCAUCGCCACAUCCGGC ...(((((.....((((.((((((........(((((.((.......)).)))))..(((((((((((........)))))))))))..))))))))))(((((....))))).))))). ( -54.00) >DroSec_CAF1 20126 120 - 1 CUCUCGGACAUGUCGUUAUUGCUGCUCACUGCGCGCCCGCCAAUGUUGCCGGCGCAUCCAGCUCCAUUAUUUGUCAAGUGGAGCUGGCCCAGCGAGGCGGUGGCCAUCGCCACAUCCGGC ...(((((.....((((.((((((........(((((.((.......)).)))))..(((((((((((........)))))))))))..))))))))))(((((....))))).))))). ( -54.00) >DroEre_CAF1 20026 120 - 1 CUCUCGGACAUGUCGUUAUUACUGCUCACUGCGCGUCCGCCAAUGUUGCCGGCGCAUCCAGCUCCAUUAUUUGUCAAGUGAAGCUGGCCCAGCGAGGCGGUGGCCAUCGCCACAUCCGGC .....((((.....((....)).((.....))..))))(((.....((((..(((..((((((.((((........)))).))))))....))).))))(((((....)))))....))) ( -42.00) >consensus CUCUCGGACAUGUCGUUAUUGCUGCUCACUGCGCGCCCGCCAAUGUUGCCGGCGCAUCCAGCUCCAUUAUUUGUCAAGUGGAGCUGGCCCAGCGAGGCGGUGGCCAUCGCCACAUCCGGC ...(((((.....((((.((((((........(((((.((.......)).)))))..(((((((((((........)))))))))))..))))))))))(((((....))))).))))). (-49.15 = -49.60 + 0.45)

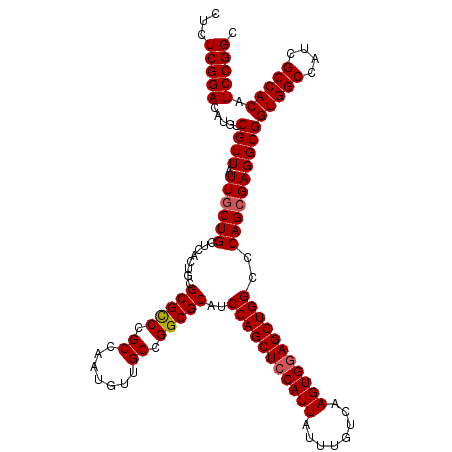

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:29 2006