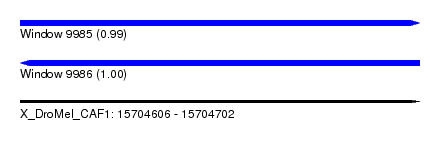
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,704,606 – 15,704,702 |
| Length | 96 |
| Max. P | 0.999986 |

| Location | 15,704,606 – 15,704,702 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 72.05 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -13.23 |
| Energy contribution | -12.97 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15704606 96 + 22224390 AUGUUUUUCAAUGC-----AAAUUGUACACAUGUAUAGAAAUUUUAAUAGAUGUUCAGUUGCACAUGUGCAACAGUGCGUUUCUCUAUACAUAUGCUCAUC- .(((........))-----)...((..((.(((((((((.........((((((...((((((....))))))...)))))).))))))))).))..))..- ( -21.50) >DroSec_CAF1 15797 96 + 1 AUGUUUUUUAAUGC-----GAAAUGUACACAUGUAUAGAAAUAUUAAUAGAUGUUCAGUUGCACAUGUGCAACAGUGCGUUUCUCUAUACAUAUGCUCAUC- ..............-----(((((((((((((.(((.((....)).))).))))...((((((....)))))).)))))))))..................- ( -22.40) >DroEre_CAF1 15598 96 + 1 AUGGAUUUUAAUAC-----GACAUGUACGCAUGUAUAGAAAUUUUAAUAGAUAUUCAGUUGCACAUGUGCAACAGUGCGAUUCUCUAUACAUAUGCCCAUC- ((((.......(((-----.....))).(((((((((((.((((....))))....((((((((.((.....)))))))))).))))))))..))))))).- ( -21.00) >DroPer_CAF1 20572 93 + 1 GUUU-CUUUAAACCAGAAGAAAU--UGCAUAUUUGUAGUCCUCGUAAUUGAGGCUCACUUGUUCGUGCGAAAGAAAG---UUCUC---AAAUAUGCUCAAGU ((((-((((......))))))))--.(((((((((.((.(((((....))))))).((((.(((....)))...)))---)...)---))))))))...... ( -23.90) >consensus AUGUUUUUUAAUGC_____AAAAUGUACACAUGUAUAGAAAUUUUAAUAGAUGUUCAGUUGCACAUGUGCAACAGUGCGUUUCUCUAUACAUAUGCUCAUC_ ...........................((.(((((((((.((((....)))).....((((((....))))))..........))))))))).))....... (-13.23 = -12.97 + -0.25)

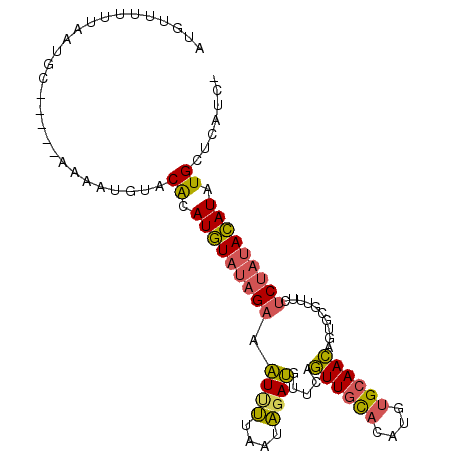
| Location | 15,704,606 – 15,704,702 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 72.05 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -17.12 |
| Energy contribution | -18.50 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 5.40 |
| SVM RNA-class probability | 0.999986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15704606 96 - 22224390 -GAUGAGCAUAUGUAUAGAGAAACGCACUGUUGCACAUGUGCAACUGAACAUCUAUUAAAAUUUCUAUACAUGUGUACAAUUU-----GCAUUGAAAAACAU -..((.(((((((((((((((........((((((....)))))).(.....)........))))))))))))))).))....-----.............. ( -25.60) >DroSec_CAF1 15797 96 - 1 -GAUGAGCAUAUGUAUAGAGAAACGCACUGUUGCACAUGUGCAACUGAACAUCUAUUAAUAUUUCUAUACAUGUGUACAUUUC-----GCAUUAAAAAACAU -((((.(((((((((((((((........((((((....)))))).(.....)........))))))))))))))).))))..-----.............. ( -27.00) >DroEre_CAF1 15598 96 - 1 -GAUGGGCAUAUGUAUAGAGAAUCGCACUGUUGCACAUGUGCAACUGAAUAUCUAUUAAAAUUUCUAUACAUGCGUACAUGUC-----GUAUUAAAAUCCAU -.(((((((..((((((((((........((((((....)))))).(.....)........)))))))))))))((((.....-----))))......)))) ( -23.20) >DroPer_CAF1 20572 93 - 1 ACUUGAGCAUAUUU---GAGAA---CUUUCUUUCGCACGAACAAGUGAGCCUCAAUUACGAGGACUACAAAUAUGCA--AUUUCUUCUGGUUUAAAG-AAAC ......((((((((---(((..---......(((((........)))))((((......)))).)).))))))))).--.((((((........)))-))). ( -22.80) >consensus _GAUGAGCAUAUGUAUAGAGAAACGCACUGUUGCACAUGUGCAACUGAACAUCUAUUAAAAUUUCUAUACAUGUGUACAAUUC_____GCAUUAAAAAACAU ......(((((((((((((((........((((((....)))))).(.....)........))))))))))))))).......................... (-17.12 = -18.50 + 1.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:18 2006