
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,702,856 – 15,703,216 |
| Length | 360 |
| Max. P | 0.965365 |

| Location | 15,702,856 – 15,702,976 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -55.00 |
| Consensus MFE | -51.50 |
| Energy contribution | -51.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15702856 120 - 22224390 GGAGUGGGCACACGGAAUGCCAUUCCGGUGAGCUUACCGUUGAGGGCGGGGAUAACCUUGCCCACGGCCUUGGCAGCUCCAGUGGAAGCUGGAAUGAUGUUCUGGGCAGCGCCACGUCCA (((((((((...((((((..((((((((((((((..((((((.((((((((....)))))))).))))...)).)))))........)))))))))..))))))....)).)))).))). ( -56.70) >DroSec_CAF1 14013 120 - 1 GCAGUGGGCACACGGAAUGCCAUUCCGGUGAGCUUACCAUUGAGGGCGGGGAUAACCUUGCCCACGGCCUUGGCAGCUCCGGUGGAAGCUGGAAUGAUGUUCUGGGCAGCGCCACGUCCA ....(((((...((((((..((((((....(((((.((((((.((((((((....))))))))...((....)).....)))))))))))))))))..))))))(((...)))..))))) ( -54.30) >DroEre_CAF1 13762 120 - 1 GGAGUGGGCACACGGAAUGCCAUUCCGGUGAGCUUACCGUUGAGGGCGGGAAUAACCUUGCCCACGGCCUUGGCAGCUCCGGUCGAAGCUGGAAUGAUGUUCUGGGCAGCGCCACGUCCA (((((((((...((((((..((((((((((((((..((((((.(((((((......))))))).))))...)).)))))........)))))))))..))))))....)).)))).))). ( -54.00) >consensus GGAGUGGGCACACGGAAUGCCAUUCCGGUGAGCUUACCGUUGAGGGCGGGGAUAACCUUGCCCACGGCCUUGGCAGCUCCGGUGGAAGCUGGAAUGAUGUUCUGGGCAGCGCCACGUCCA (((((((((...((((((..((((((((((((((.........(((((((......)))))))...((....)))))))........)))))))))..))))))....)).)))).))). (-51.50 = -51.83 + 0.33)

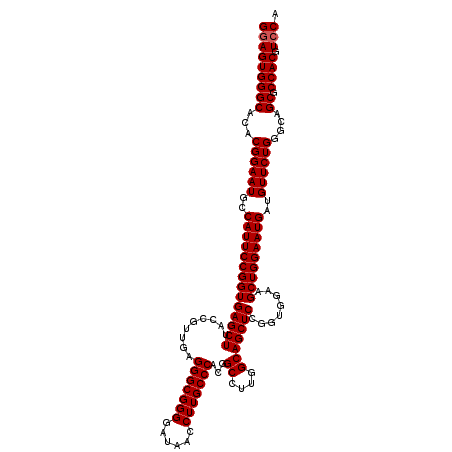
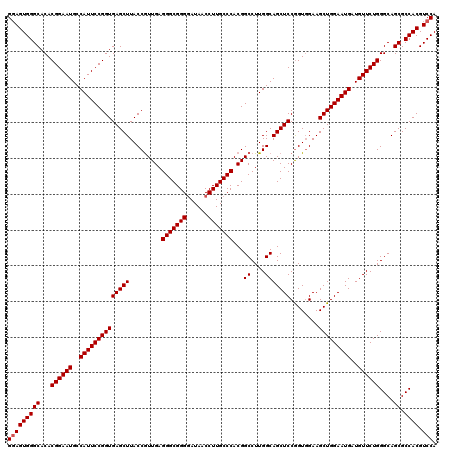
| Location | 15,702,896 – 15,703,016 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -54.50 |
| Consensus MFE | -51.17 |
| Energy contribution | -51.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15702896 120 - 22224390 CUUGCCCAAGCGCACGGUCAAAUCGACCACGGAAACGUUGGGAGUGGGCACACGGAAUGCCAUUCCGGUGAGCUUACCGUUGAGGGCGGGGAUAACCUUGCCCACGGCCUUGGCAGCUCC ..((((((..(.((.((((.....))))(((....))))).)..))))))...(((.(((((....((((....))))((((.((((((((....)))))))).))))..)))))..))) ( -56.00) >DroSec_CAF1 14053 120 - 1 CUUGCCCAAGCGCACGGUCAAAUCGACCACGGAAACGUUGGCAGUGGGCACACGGAAUGCCAUUCCGGUGAGCUUACCAUUGAGGGCGGGGAUAACCUUGCCCACGGCCUUGGCAGCUCC ..((((((...((..((((.....))))(((....)))..))..))))))...(((.(((((..((((((....)))).....((((((((....))))))))..))...)))))..))) ( -54.20) >DroEre_CAF1 13802 120 - 1 CUUGCCCAAGCGCACGGUCAGAUCGACCACGGAAACGUUGGGAGUGGGCACACGGAAUGCCAUUCCGGUGAGCUUACCGUUGAGGGCGGGAAUAACCUUGCCCACGGCCUUGGCAGCUCC ..((((((..(.((.((((.....))))(((....))))).)..))))))...(((.(((((....((((....))))((((.(((((((......))))))).))))..)))))..))) ( -53.30) >consensus CUUGCCCAAGCGCACGGUCAAAUCGACCACGGAAACGUUGGGAGUGGGCACACGGAAUGCCAUUCCGGUGAGCUUACCGUUGAGGGCGGGGAUAACCUUGCCCACGGCCUUGGCAGCUCC ..((((((..(.((.((((.....))))(((....))))).)..))))))...(((.(((((..((((((....)))).....(((((((......)))))))..))...)))))..))) (-51.17 = -51.50 + 0.33)

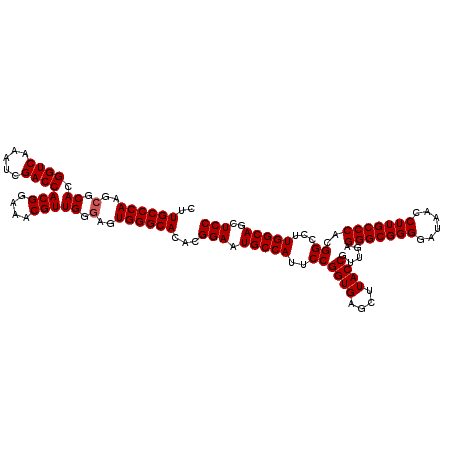

| Location | 15,703,016 – 15,703,136 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -46.03 |
| Consensus MFE | -44.31 |
| Energy contribution | -44.20 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15703016 120 + 22224390 GGUGCGUCCUAUGAUGAAAUUAAGGCCAAGGUUCAGGAGGCCGCCAACGGACCCCUGAAGGGUAUCCUGGGAUACACCGAUGAGGAGGUCGUUUCUACCGAUUUCCUCAGCGACACCCAC ((((..(((((.(((.........(((....((((((.(((((....))).)))))))).)))))).)))))..))))(.(((((((((((.......))))))))))).)......... ( -48.10) >DroSec_CAF1 14173 120 + 1 GGUGCGUCCUAUGAUGAAAUUAAGGCCAAGGUUGAGGAGGCCGCCAACGGACCCCUGAAGGGAAUCCUGGGAUACACCGAUGAGGAGGUCGUGUCCACCGAUUUCCUCAGCGACACCCAC ((((..(((((.(((........(((...((((.....))))))).......(((....))).))).)))))..))))(.(((((((((((.(....)))))))))))).)......... ( -44.20) >DroEre_CAF1 13922 120 + 1 GGUGCAUCCUAUGAUGAAAUUAAGGCCAAGGUUCAGGAGGCCGCCAACGGACCCCUGAAGGGAAUUUUGGGAUACACCGAUGAGGAGGUCGUGUCCACCGAUUUCCUCAGCGACACCCAC ((((.((((((.(((..........((....((((((.(((((....))).)))))))).)).))).)))))).))))(.(((((((((((.(....)))))))))))).)......... ( -45.80) >consensus GGUGCGUCCUAUGAUGAAAUUAAGGCCAAGGUUCAGGAGGCCGCCAACGGACCCCUGAAGGGAAUCCUGGGAUACACCGAUGAGGAGGUCGUGUCCACCGAUUUCCUCAGCGACACCCAC ((((.((((((.(((..........((....((((((.(((((....))).)))))))).)).))).)))))).))))(.(((((((((((.......))))))))))).)......... (-44.31 = -44.20 + -0.11)


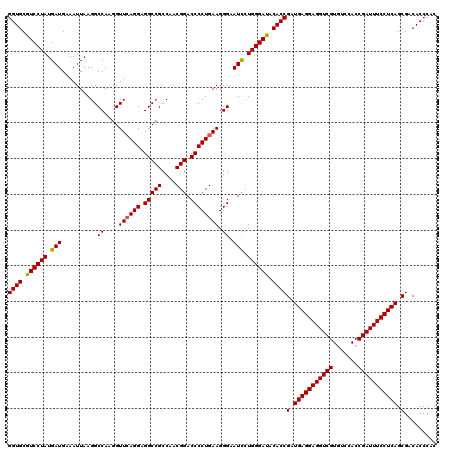
| Location | 15,703,096 – 15,703,216 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -44.50 |
| Consensus MFE | -41.70 |
| Energy contribution | -41.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15703096 120 + 22224390 UGAGGAGGUCGUUUCUACCGAUUUCCUCAGCGACACCCACUCGUCGGUGUUCGAUGCCAAGGCUGGCAUUUCGCUAAACGACAAGUUCGUGAAGCUGAUCUCUUGGUACGACAACGAGUU .((((((((((.......))))))))))(((..(((..(((.((((..((..(((((((....)))))))..))....)))).)))..)))..)))...((((((......))).))).. ( -43.00) >DroSec_CAF1 14253 120 + 1 UGAGGAGGUCGUGUCCACCGAUUUCCUCAGCGACACCCACUCGUCGGUGUUCGAUGCCAAGGCUGGCAUUUCGCUGAACGACAAGUUCGUGAAGCUGAUCUCUUGGUACGACAACGAGUU (((((((((((.(....)))))))))))).........((((((...(((.((.((((((((....((((((((.((((.....)))))))))).))...))))))))))))))))))). ( -45.70) >DroEre_CAF1 14002 120 + 1 UGAGGAGGUCGUGUCCACCGAUUUCCUCAGCGACACCCACUCGUCGGUCUUCGAUGCCAAGGCUGGCAUUUCGCUGAACGACAAGUUCGUGAAGCUGAUCUCUUGGUACGACAACGAGUU .((((((((((.(....)))))))))))(((..(((..(((.((((.((..((((((((....)))))..)))..)).)))).)))..)))..)))...((((((......))).))).. ( -44.80) >consensus UGAGGAGGUCGUGUCCACCGAUUUCCUCAGCGACACCCACUCGUCGGUGUUCGAUGCCAAGGCUGGCAUUUCGCUGAACGACAAGUUCGUGAAGCUGAUCUCUUGGUACGACAACGAGUU .((((((((((.......))))))))))(((..(((..(((.((((.....((((((((....)))))..))).....)))).)))..)))..)))...((((((......))).))).. (-41.70 = -41.70 + -0.00)


| Location | 15,703,096 – 15,703,216 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -43.98 |
| Consensus MFE | -42.47 |
| Energy contribution | -42.59 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15703096 120 - 22224390 AACUCGUUGUCGUACCAAGAGAUCAGCUUCACGAACUUGUCGUUUAGCGAAAUGCCAGCCUUGGCAUCGAACACCGACGAGUGGGUGUCGCUGAGGAAAUCGGUAGAAACGACCUCCUCA ..(((.(((......))))))...(((..(((..((((((((.....(((..((((......))))))).....))))))))..)))..)))(((((..(((.......)))..))))). ( -41.80) >DroSec_CAF1 14253 120 - 1 AACUCGUUGUCGUACCAAGAGAUCAGCUUCACGAACUUGUCGUUCAGCGAAAUGCCAGCCUUGGCAUCGAACACCGACGAGUGGGUGUCGCUGAGGAAAUCGGUGGACACGACCUCCUCA ...((((.(((.((((......(((((..(((..((((((((.....(((..((((......))))))).....))))))))..)))..))))).......)))))))))))........ ( -43.92) >DroEre_CAF1 14002 120 - 1 AACUCGUUGUCGUACCAAGAGAUCAGCUUCACGAACUUGUCGUUCAGCGAAAUGCCAGCCUUGGCAUCGAAGACCGACGAGUGGGUGUCGCUGAGGAAAUCGGUGGACACGACCUCCUCA ...((((.(((.((((......(((((..(((..((((((((.((..(((..((((......)))))))..)).))))))))..)))..))))).......)))))))))))........ ( -46.22) >consensus AACUCGUUGUCGUACCAAGAGAUCAGCUUCACGAACUUGUCGUUCAGCGAAAUGCCAGCCUUGGCAUCGAACACCGACGAGUGGGUGUCGCUGAGGAAAUCGGUGGACACGACCUCCUCA ...((((.(((.((((......(((((..(((..((((((((.....(((..((((......))))))).....))))))))..)))..))))).......)))))))))))........ (-42.47 = -42.59 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:16 2006