
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,692,897 – 15,693,097 |
| Length | 200 |
| Max. P | 0.992454 |

| Location | 15,692,897 – 15,693,017 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -30.82 |
| Energy contribution | -31.74 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15692897 120 - 22224390 AAUCGCCAAAAGAUUGGCGAACCUAUACAUUAACUAACCAUCCCGGCAUUAUUGUGAUUCGGAAUCCAUUCUCGGAACGAGGCCGGCGUUAUUGGAGCGCCCGAUGUCUGCGGGAUUUUC ..(((((((....)))))))...................((((((((....((((..(((((((....))).)))))))).)))((((((.....))))))..........))))).... ( -36.20) >DroSec_CAF1 3941 120 - 1 AAUCGCCAAAAGAUUGGCGAUCCUAUACAUUAACUAACCAUCCCGGCAUUAUUGUGAUUCGGAAUCCAUUCUCGGAACGUGGCCGGCGUUAUUGGAUCGCCCGAUGUCUGCGGGAUUAUC .((((((((....))))))))..................(((((((((((...(((((((((..(((......)))((((.....))))..)))))))))..)))))...)))))).... ( -35.10) >DroSim_CAF1 3946 120 - 1 AAUCGCCAAAAGAUUGGCGAUCCUAUACAUUAACUAACCAUCCCGGCAUUAUUGUGAUUCGGAAUCCAUUCUCGGAACGUGGCCGGCGUUAUUGGAUCGCCCGAUGUCUGCGGGAUUAUC .((((((((....))))))))..................(((((((((((...(((((((((..(((......)))((((.....))))..)))))))))..)))))...)))))).... ( -35.10) >DroEre_CAF1 3934 120 - 1 AAUCGCCAAAAGAUUGGCGAUCUUAUACAUUAACUAAUCAUCCCGGCAUUAUUGUUAUUCGGAACCCAUUCUCGGAACGAGGACGACGUUAUUGGAUAGCCCGAUGUCUGCGGGAUUAUC .((((((((....))))))))..................(((((((((((...((((((((((((.(.((((((...)))))).)..))).)))))))))..)))))...)))))).... ( -34.40) >DroYak_CAF1 3873 120 - 1 UAUCGCCUAAGGAUUGGCGAUCUUAUACAUUACCUAAUCAUCCCGGCAUUAUUGUUAUUCGGAACCCAUUCUCGGAACGAGGUCGGCGUUAUUGGAUCGCCCGAUGUCUGCGGGAUUAUC .((((((........))))))..................(((((((((((...((.(((((((((((..(((((...)))))..)).))).)))))).))..)))))...)))))).... ( -32.90) >consensus AAUCGCCAAAAGAUUGGCGAUCCUAUACAUUAACUAACCAUCCCGGCAUUAUUGUGAUUCGGAAUCCAUUCUCGGAACGAGGCCGGCGUUAUUGGAUCGCCCGAUGUCUGCGGGAUUAUC .((((((((....))))))))..................(((((((((((...((((((((((((((..(((((...)))))..)).))).)))))))))..)))))...)))))).... (-30.82 = -31.74 + 0.92)

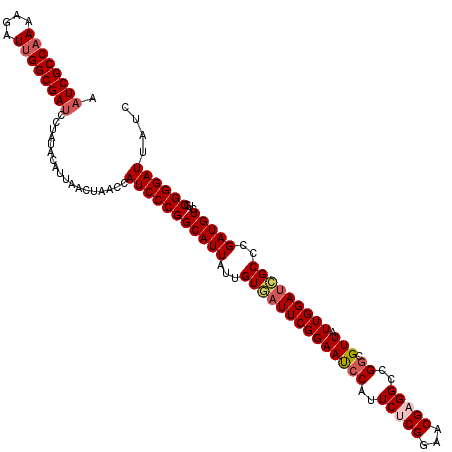

| Location | 15,692,937 – 15,693,057 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -30.86 |
| Energy contribution | -30.78 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15692937 120 + 22224390 UCGUUCCGAGAAUGGAUUCCGAAUCACAAUAAUGCCGGGAUGGUUAGUUAAUGUAUAGGUUCGCCAAUCUUUUGGCGAUUUAACGCCUGGCAUUGUCUGUAGGCUUUCUACAUUGCUGAU (((.(((......)))...)))(((((((((((((((((...(((((.............(((((((....))))))).))))).))))))))))).(((((.....))))).)).)))) ( -36.54) >DroSec_CAF1 3981 120 + 1 ACGUUCCGAGAAUGGAUUCCGAAUCACAAUAAUGCCGGGAUGGUUAGUUAAUGUAUAGGAUCGCCAAUCUUUUGGCGAUUUAACGCCUGGCAUUGUCUGGGGGCUUUCUACAUUGCUGAU ..(((((.(((.(((((.....))).)).((((((((((...((((............(((((((((....))))))))))))).))))))))))))).)))))................ ( -39.20) >DroSim_CAF1 3986 120 + 1 ACGUUCCGAGAAUGGAUUCCGAAUCACAAUAAUGCCGGGAUGGUUAGUUAAUGUAUAGGAUCGCCAAUCUUUUGGCGAUUUAACGCCUGGCAUUGUCUGUGGGCUUUCUACAUUGCUGAU .((.(((......)))...))...(((((((((((((((...((((............(((((((((....))))))))))))).))))))))))).))))(((..........)))... ( -38.30) >DroEre_CAF1 3974 120 + 1 UCGUUCCGAGAAUGGGUUCCGAAUAACAAUAAUGCCGGGAUGAUUAGUUAAUGUAUAAGAUCGCCAAUCUUUUGGCGAUUUUAUGCCUGGCAUUGUCUGUAAGCUUUCUACAUUGCUGAA (((.(((......)))...)))...((((((((((((((((.....)))...(((((((((((((((....))))))))))))))))))))))))).))).(((..........)))... ( -39.10) >DroYak_CAF1 3913 120 + 1 UCGUUCCGAGAAUGGGUUCCGAAUAACAAUAAUGCCGGGAUGAUUAGGUAAUGUAUAAGAUCGCCAAUCCUUAGGCGAUAUAAUGCCUGGCAUUGUCUGAAGGCUUUCUACAUUGCUGAU ((((((((.(.((..(((......)))....)).)))))))))...(((((((((.(((((((((........)))))).....((((..((.....)).))))))).)))))))))... ( -33.50) >consensus UCGUUCCGAGAAUGGAUUCCGAAUCACAAUAAUGCCGGGAUGGUUAGUUAAUGUAUAGGAUCGCCAAUCUUUUGGCGAUUUAACGCCUGGCAUUGUCUGUAGGCUUUCUACAUUGCUGAU .((.(((......)))...)).....(((((((((((((...((((............(((((((((....))))))))))))).))))))))))).))..(((..........)))... (-30.86 = -30.78 + -0.08)

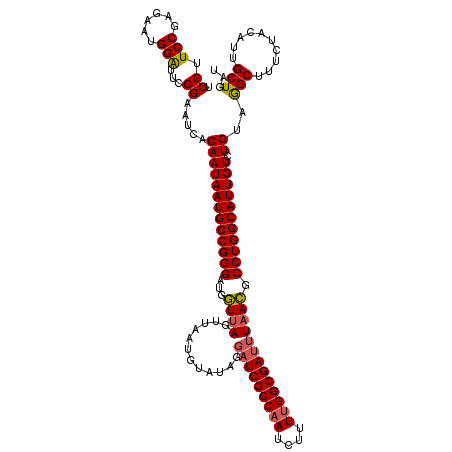

| Location | 15,692,977 – 15,693,097 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.75 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.26 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15692977 120 + 22224390 UGGUUAGUUAAUGUAUAGGUUCGCCAAUCUUUUGGCGAUUUAACGCCUGGCAUUGUCUGUAGGCUUUCUACAUUGCUGAUACUUAAUCGUUGAACAAUAUUCUGAGGGGAAUGUAGAAAU ....(((.((((((.((((((((((((....)))))))......))))))))))).))).....((((((((((.(((((.....)))...(((.....)))....)).)))))))))). ( -32.40) >DroSec_CAF1 4021 120 + 1 UGGUUAGUUAAUGUAUAGGAUCGCCAAUCUUUUGGCGAUUUAACGCCUGGCAUUGUCUGGGGGCUUUCUACAUUGCUGAUACUUAAUCGUUGAACAAUAUUCUGAGGGAAAUGUAUAAAU ..((((((.((((((..((((((((((....))))))))))..(.((..(......)..)).).....)))))))))))).........(((.(((...(((.....))).))).))).. ( -28.30) >DroSim_CAF1 4026 120 + 1 UGGUUAGUUAAUGUAUAGGAUCGCCAAUCUUUUGGCGAUUUAACGCCUGGCAUUGUCUGUGGGCUUUCUACAUUGCUGAUACUUAAUCGUUGAACAAUAUUCUGAGGGAAAUGUAAAAAU ..((((((.((((((..((((((((((....))))))))))...(((((.((.....)))))))....)))))))))))).............(((...(((.....))).)))...... ( -28.40) >DroEre_CAF1 4014 120 + 1 UGAUUAGUUAAUGUAUAAGAUCGCCAAUCUUUUGGCGAUUUUAUGCCUGGCAUUGUCUGUAAGCUUUCUACAUUGCUGAACAAUAUUCGUUGAACAAUAUUCUAAUGUAAAUGUAGAAAU .(((..((((..(((((((((((((((....))))))))))))))).))))...))).......((((((((((((.....((((((.(.....))))))).....)).)))))))))). ( -34.70) >DroYak_CAF1 3953 120 + 1 UGAUUAGGUAAUGUAUAAGAUCGCCAAUCCUUAGGCGAUAUAAUGCCUGGCAUUGUCUGAAGGCUUUCUACAUUGCUGAUACAUAAUCGCUGAACAACAUUCUAAAGUAAAUGUUGAAAU .((((((((((((((.(((((((((........)))))).....((((..((.....)).))))))).)))))))))......)))))......(((((((........))))))).... ( -31.00) >consensus UGGUUAGUUAAUGUAUAGGAUCGCCAAUCUUUUGGCGAUUUAACGCCUGGCAUUGUCUGUAGGCUUUCUACAUUGCUGAUACUUAAUCGUUGAACAAUAUUCUGAGGGAAAUGUAGAAAU ..(((((.(((((((..((((((((((....))))))))))...(((((((...)))...))))....))))))))))))................(((((........)))))...... (-24.02 = -24.26 + 0.24)

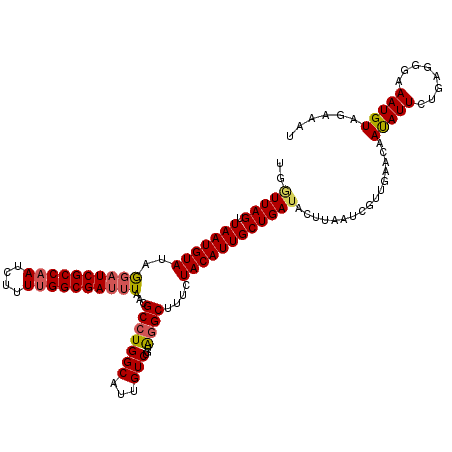
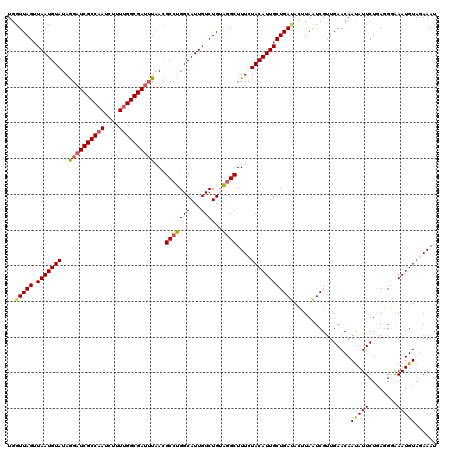
| Location | 15,692,977 – 15,693,097 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.75 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15692977 120 - 22224390 AUUUCUACAUUCCCCUCAGAAUAUUGUUCAACGAUUAAGUAUCAGCAAUGUAGAAAGCCUACAGACAAUGCCAGGCGUUAAAUCGCCAAAAGAUUGGCGAACCUAUACAUUAACUAACCA .((((((((((.(.....((((...))))...(((.....))).).))))))))))((((.((.....))..))))......(((((((....))))))).................... ( -26.50) >DroSec_CAF1 4021 120 - 1 AUUUAUACAUUUCCCUCAGAAUAUUGUUCAACGAUUAAGUAUCAGCAAUGUAGAAAGCCCCCAGACAAUGCCAGGCGUUAAAUCGCCAAAAGAUUGGCGAUCCUAUACAUUAACUAACCA ....................((((((((..((......))...)))))))).....(((..((.....))...)))(((((((((((((....))))))))........)))))...... ( -20.70) >DroSim_CAF1 4026 120 - 1 AUUUUUACAUUUCCCUCAGAAUAUUGUUCAACGAUUAAGUAUCAGCAAUGUAGAAAGCCCACAGACAAUGCCAGGCGUUAAAUCGCCAAAAGAUUGGCGAUCCUAUACAUUAACUAACCA .((((((((((.(.....((((...))))...(((.....))).).))))))))))(((..((.....))...)))(((((((((((((....))))))))........)))))...... ( -23.60) >DroEre_CAF1 4014 120 - 1 AUUUCUACAUUUACAUUAGAAUAUUGUUCAACGAAUAUUGUUCAGCAAUGUAGAAAGCUUACAGACAAUGCCAGGCAUAAAAUCGCCAAAAGAUUGGCGAUCUUAUACAUUAACUAAUCA .((((((((((.......(((((.(((((...))))).)))))...))))))))))((((.((.....))..))))((((.((((((((....)))))))).)))).............. ( -29.30) >DroYak_CAF1 3953 120 - 1 AUUUCAACAUUUACUUUAGAAUGUUGUUCAGCGAUUAUGUAUCAGCAAUGUAGAAAGCCUUCAGACAAUGCCAGGCAUUAUAUCGCCUAAGGAUUGGCGAUCUUAUACAUUACCUAAUCA ....((((((((......))))))))......(((((.(((......(((((....((((.((.....))..)))).....((((((........))))))....)))))))).))))). ( -27.90) >consensus AUUUCUACAUUUCCCUCAGAAUAUUGUUCAACGAUUAAGUAUCAGCAAUGUAGAAAGCCUACAGACAAUGCCAGGCGUUAAAUCGCCAAAAGAUUGGCGAUCCUAUACAUUAACUAACCA .((((((((((.......((((...))))...(((.....)))...))))))))))(((..((.....))...))).....((((((((....))))))))................... (-21.32 = -21.52 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:04 2006