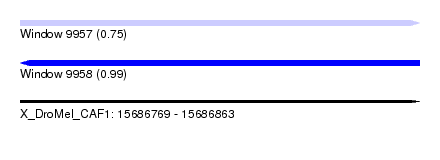
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,686,769 – 15,686,863 |
| Length | 94 |
| Max. P | 0.993045 |

| Location | 15,686,769 – 15,686,863 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 84.41 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -10.87 |
| Energy contribution | -11.03 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15686769 94 + 22224390 UUGAAUUUUUCUCAAUUUUAUAUCAAUGCCUUUGUACACACACAUACGCAUAUGCAUAUAUGCAUAUGUACGUAUGUAUGUGCUUGUGUAGUUG ((((.......))))...................((((((((((((((((((((((....)))))))))......)))))))..)))))).... ( -23.70) >DroSec_CAF1 2444 86 + 1 UUGAAUUUUUCUCAAUUUUAUAUCAAUGCCUUUGUACACACACAUACGCAUAUG--------CAUAUGUACGUAUGUAUGUGUUUGUGUAGUUG ((((.......))))...................((((((.(((((..((((((--------........))))))..))))).)))))).... ( -19.60) >DroSim_CAF1 2319 86 + 1 UUGAAUUUUUCUCAAUUUUAUAUCAAUGCCUUUGUACACACACAUACGCAUAUG--------CAUAUGUACGUAUGUAUGUGUUUGUGUAGUUG ((((.......))))...................((((((.(((((..((((((--------........))))))..))))).)))))).... ( -19.60) >DroEre_CAF1 3776 83 + 1 UUGAAUUUUUCUCAAUUUUAUUUCAAUGCCUUUGUAAAUACACAUAUGCAUAUGCA----------CGUAUGUAUGUAUGU-CGUGUGUAGUUG (((((................)))))...........((((((((((((((((...----------.....))))))))))-.))))))..... ( -16.19) >DroYak_CAF1 2322 84 + 1 UUGAAUUUUUCUCAACUUUAAUUCAAUGCCUUUGUACAUACACACAUGCAUAUGUA----------CGUUUGCAUGUGUGUGCGUGUGCAGUUG (((((((............))))))).....(((((((((((((((((((.((...----------.)).)))))))))))..))))))))... ( -28.00) >consensus UUGAAUUUUUCUCAAUUUUAUAUCAAUGCCUUUGUACACACACAUACGCAUAUG_A______CAUAUGUACGUAUGUAUGUGCUUGUGUAGUUG ((((.......))))...................((((((..((((..((((((................))))))..))))..)))))).... (-10.87 = -11.03 + 0.16)

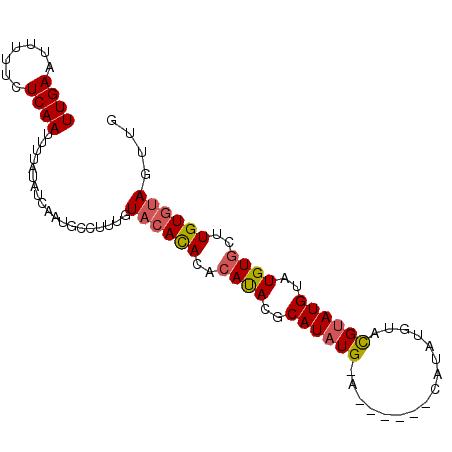

| Location | 15,686,769 – 15,686,863 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.41 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -13.64 |
| Energy contribution | -15.68 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15686769 94 - 22224390 CAACUACACAAGCACAUACAUACGUACAUAUGCAUAUAUGCAUAUGCGUAUGUGUGUGUACAAAGGCAUUGAUAUAAAAUUGAGAAAAAUUCAA ...........(((((((((((((..((((((((....)))))))))))))))))))))................................... ( -28.20) >DroSec_CAF1 2444 86 - 1 CAACUACACAAACACAUACAUACGUACAUAUG--------CAUAUGCGUAUGUGUGUGUACAAAGGCAUUGAUAUAAAAUUGAGAAAAAUUCAA ...........(((((((((((((((.((...--------.)).)))))))))))))))................................... ( -20.40) >DroSim_CAF1 2319 86 - 1 CAACUACACAAACACAUACAUACGUACAUAUG--------CAUAUGCGUAUGUGUGUGUACAAAGGCAUUGAUAUAAAAUUGAGAAAAAUUCAA ...........(((((((((((((((.((...--------.)).)))))))))))))))................................... ( -20.40) >DroEre_CAF1 3776 83 - 1 CAACUACACACG-ACAUACAUACAUACG----------UGCAUAUGCAUAUGUGUAUUUACAAAGGCAUUGAAAUAAAAUUGAGAAAAAUUCAA ............-......(((((((..----------(((....)))..)))))))..................................... ( -10.60) >DroYak_CAF1 2322 84 - 1 CAACUGCACACGCACACACAUGCAAACG----------UACAUAUGCAUGUGUGUAUGUACAAAGGCAUUGAAUUAAAGUUGAGAAAAAUUCAA (((((((....))(((((((((((....----------......)))))))))))((((......))))........)))))............ ( -21.50) >consensus CAACUACACAAGCACAUACAUACGUACAUAUG______U_CAUAUGCGUAUGUGUGUGUACAAAGGCAUUGAUAUAAAAUUGAGAAAAAUUCAA ...........(((((((((((((..((((((........)))))))))))))))))))................................... (-13.64 = -15.68 + 2.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:52 2006