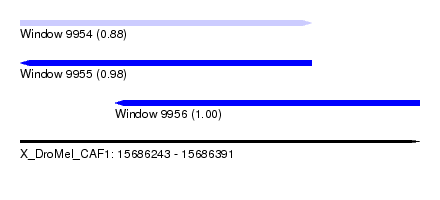
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,686,243 – 15,686,391 |
| Length | 148 |
| Max. P | 0.999836 |

| Location | 15,686,243 – 15,686,351 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -22.71 |
| Energy contribution | -22.31 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15686243 108 + 22224390 --UCGUAUUUGUAUUUUG--UGUCUUCAAGUGAUUCAGU--------AAUUCACUGACUUAACUGCCCAUCCGAAUUCGAAAUGGGCAAGUUUCCAGGUCGCAUGUAAUCAGCACAAUCA --.............(((--(((...((.((((((((((--------.....)))))((.((((((((((.((....))..)))))).))))...))))))).))......))))))... ( -27.00) >DroSec_CAF1 1924 110 + 1 --UCGUAUUUGUAUUUUGUGUGUCUUCAAGUGAUUCAAU--------AAUUCACUGACUUAACUGCCCAUCCGAUUUCGAAAUGGGCAAGUUUCCAGGUCGUAUGUAAUCAGCACAAUCA --.............((((((.....(((((((......--------...)))))(((((((((((((((.((....))..)))))).))))...)))))...))......))))))... ( -27.50) >DroSim_CAF1 1799 110 + 1 --UCGUAUUUGUAUUUUGUGUGUCUUCAAGUGAUUCAAU--------AAUUCACUGACUUAACUGCCCAUCCGAUUUCGAAAUGGGCAAGUUUCCAGGUCGUAUGUAAUCAGCACAAUCA --.............((((((.....(((((((......--------...)))))(((((((((((((((.((....))..)))))).))))...)))))...))......))))))... ( -27.50) >DroEre_CAF1 3285 116 + 1 --UCGUAUUUGUAUUUUGUGCU--UCCAAGUGGUUCAGUAAUGAAGUAAUUCACUGACUUAACUGCCCAUCCGAUUUCGAAAUGGGCAAGUUUCCAGGUCGUAUGUAAUCAGUGCAUUCA --..(((((.(.(((.(((((.--....(((((((((....)))).....)))))(((((((((((((((.((....))..)))))).))))...)))))))))).)))))))))..... ( -29.30) >DroYak_CAF1 1789 120 + 1 UUUUGUAUUUGUAUUCUGUGUUUUUUCAAGUGAUUCAGUAAUGCAGUAAUUCACUGAUUUAACUGCCCAUCCGAUUUCGAAAUGGGCAAGUUUCCAGGUCGUAUGUAAUCAGUGCAAUCA ........((((((((((.(((.........))).))).))))))).....((((((((.((((((((((.((....))..)))))).))))..............))))))))...... ( -27.26) >consensus __UCGUAUUUGUAUUUUGUGUGUCUUCAAGUGAUUCAGU________AAUUCACUGACUUAACUGCCCAUCCGAUUUCGAAAUGGGCAAGUUUCCAGGUCGUAUGUAAUCAGCACAAUCA ................(((((.......(((((.................)))))(((((((((((((((.((....))..)))))).))))...)))))...........))))).... (-22.71 = -22.31 + -0.40)

| Location | 15,686,243 – 15,686,351 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -24.00 |
| Energy contribution | -23.36 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15686243 108 - 22224390 UGAUUGUGCUGAUUACAUGCGACCUGGAAACUUGCCCAUUUCGAAUUCGGAUGGGCAGUUAAGUCAGUGAAUU--------ACUGAAUCACUUGAAGACA--CAAAAUACAAAUACGA-- ...(((((((..........(((..(....)(((((((((.((....)))))))))))....)))(((((...--------......)))))...)).))--))).............-- ( -26.10) >DroSec_CAF1 1924 110 - 1 UGAUUGUGCUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAGUCAGUGAAUU--------AUUGAAUCACUUGAAGACACACAAAAUACAAAUACGA-- ...(((((.((.........(((..(....)(((((((((.((....)))))))))))....)))(((((...--------......)))))......))))))).............-- ( -26.00) >DroSim_CAF1 1799 110 - 1 UGAUUGUGCUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAGUCAGUGAAUU--------AUUGAAUCACUUGAAGACACACAAAAUACAAAUACGA-- ...(((((.((.........(((..(....)(((((((((.((....)))))))))))....)))(((((...--------......)))))......))))))).............-- ( -26.00) >DroEre_CAF1 3285 116 - 1 UGAAUGCACUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAGUCAGUGAAUUACUUCAUUACUGAACCACUUGGA--AGCACAAAAUACAAAUACGA-- ..(((.((((((((...........(....)(((((((((.((....)))))))))))...)))))))).)))..((((....))))((....)).--....................-- ( -26.60) >DroYak_CAF1 1789 120 - 1 UGAUUGCACUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAAUCAGUGAAUUACUGCAUUACUGAAUCACUUGAAAAAACACAGAAUACAAAUACAAAA (((((.((((((((...........(....)(((((((((.((....)))))))))))...))))))))))))).((.(((.(((.................)))))).))......... ( -26.93) >consensus UGAUUGUGCUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAGUCAGUGAAUU________ACUGAAUCACUUGAAGACACACAAAAUACAAAUACGA__ ......((((((((...........(....)(((((((((.((....)))))))))))...))))))))................................................... (-24.00 = -23.36 + -0.64)


| Location | 15,686,278 – 15,686,391 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.79 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -33.88 |
| Energy contribution | -32.84 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.10 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.21 |
| SVM RNA-class probability | 0.999836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15686278 113 - 22224390 GCUCAAUGUUCCAUGGAAAUAGCAAAAUGGCAUUGGCAAUUGAUUGUGCUGAUUACAUGCGACCUGGAAACUUGCCCAUUUCGAAUUCGGAUGGGCAGUUAAGUCAGUGAAUU------- ((.(((((..((((............)))))))))))..........(((((((...........(....)(((((((((.((....)))))))))))...))))))).....------- ( -31.60) >DroSec_CAF1 1961 113 - 1 GCUCGAUGCUCCAUGGAAAUAGCAAAAUGGCAUUGGCAAUUGAUUGUGCUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAGUCAGUGAAUU------- ((.(((((((.....(......).....)))))))))..........(((((((...........(....)(((((((((.((....)))))))))))...))))))).....------- ( -32.20) >DroSim_CAF1 1836 113 - 1 GCUCGAUGCUCCAUGGAAAUAGCAAAAUGGCAUUGGCAAUUGAUUGUGCUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAGUCAGUGAAUU------- ((.(((((((.....(......).....)))))))))..........(((((((...........(....)(((((((((.((....)))))))))))...))))))).....------- ( -32.20) >DroEre_CAF1 3321 120 - 1 GCCCAAUGCUCCAUGGAAAUAGCAAAAUGGCAUUGGCAAUUGAAUGCACUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAGUCAGUGAAUUACUUCAU (((.((((((.....(......).....)))))))))...((((..((((((((...........(....)(((((((((.((....)))))))))))...))))))))......)))). ( -35.70) >DroYak_CAF1 1829 120 - 1 GCUCAAUGCUCCAUGGAAAUAGCAAAAUGGCAUUGGCAAUUGAUUGCACUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAAUCAGUGAAUUACUGCAU ...(((((((.....(......).....)))))))(((..(((((.((((((((...........(....)(((((((((.((....)))))))))))...))))))))))))).))).. ( -37.10) >consensus GCUCAAUGCUCCAUGGAAAUAGCAAAAUGGCAUUGGCAAUUGAUUGUGCUGAUUACAUACGACCUGGAAACUUGCCCAUUUCGAAAUCGGAUGGGCAGUUAAGUCAGUGAAUU_______ ((.(((((((.....(......).....))))))))).........((((((((...........(....)(((((((((.((....)))))))))))...))))))))........... (-33.88 = -32.84 + -1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:50 2006