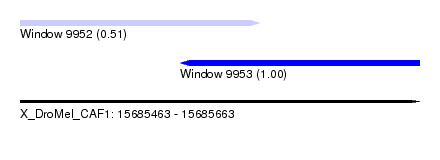
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,685,463 – 15,685,663 |
| Length | 200 |
| Max. P | 0.995970 |

| Location | 15,685,463 – 15,685,583 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -24.86 |
| Energy contribution | -26.62 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15685463 120 + 22224390 CAACUAUUCAAUAGCCAGCCACCCAGUGUGCUCAAUAAAUGCCUAAAAUAACACUUACUUUAUCAGAUGGUAUUGUUCGGUGCAUUUUCUGUGUGGUGGUGGCCAAUGGGGUGGUGAUAU ..(((((((....((((.((((((((.((((.(....((((((....((((........)))).....)))))).....).))))...)))...))))))))).....)))))))..... ( -34.10) >DroSec_CAF1 1135 120 + 1 CAACUAUUCAAUAGCCAGCCACCCACUGUGCUCAAUAAAUGCCUAAAAUAACACUUACUUUAUCAGAUGGUAUUGUUCGGUGCAUUUUCUGUGUGGUGGUGGCCAAUGGGGUGGUGAUAU ..(((((((....((((.((((((((.((((.(....((((((....((((........)))).....)))))).....).)))).....))).))))))))).....)))))))..... ( -35.60) >DroSim_CAF1 1013 120 + 1 CAACUAUUCAAUAGCCAGCCACCCACUGUGCUCAAUAAAUGCCUAAAAUAACACUUACUUUAUCAGAUGGUAUUGUUCGGUGCAUUUUCUGUGUGGUGGUGGCCAACGGGGUGGUGAUAU ..(((((((....((((.((((((((.((((.(....((((((....((((........)))).....)))))).....).)))).....))).))))))))).....)))))))..... ( -35.60) >DroEre_CAF1 2551 116 + 1 CAAGCAUUCAAUAUC----CACCCGGUGUACUCAAUAAAUGCCUAAUAUAAUACUUACUUUAUCAGAUGGUAUUGUUCGGUGCAUUUUCUGUGUAGUGGAGGCCAAAGGGGUGGUGAUAU ..............(----(((((..((..(((....((((((....((((........)))).....))))))...((.(((((.....))))).)))))..))...))))))...... ( -28.10) >DroYak_CAF1 1027 116 + 1 CAAGCAUUCAAUAGC----CACCCACUGUACUCAAUAAAUGCCUAAAAUAAUACUUACUUUAUCAGAUGGUAUUGUUCGGUGCAUUUUGUGUGUGGUGGUGGCCAAUGGGGUGGUGAUCU ((..(((((....((----((((.((..(((.(((..(((((((..(((((((((..((.....))..))))))))).)).)))))))).)))..)))))))).....))))).)).... ( -36.40) >consensus CAACUAUUCAAUAGCCAGCCACCCACUGUGCUCAAUAAAUGCCUAAAAUAACACUUACUUUAUCAGAUGGUAUUGUUCGGUGCAUUUUCUGUGUGGUGGUGGCCAAUGGGGUGGUGAUAU ..(((((((....((((.((((((((((((((.(((((.((((....((((........)))).....))))))))).))))))......))).))))))))).....)))))))..... (-24.86 = -26.62 + 1.76)



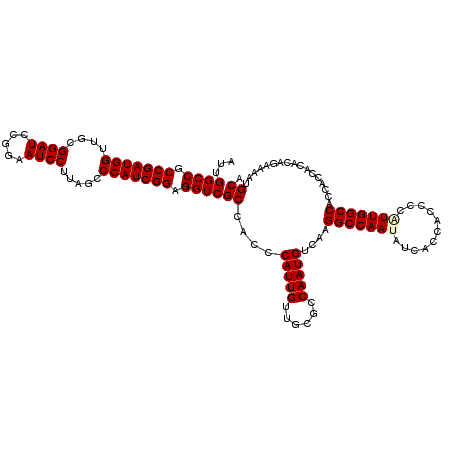
| Location | 15,685,543 – 15,685,663 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -39.01 |
| Consensus MFE | -35.28 |
| Energy contribution | -35.60 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15685543 120 - 22224390 AUUGGCCGCCGAUGGUUGCGGAUCCGGAAUCCUUAGCCCAUCGGAGGUCGCCACCCAUUGUUGCGCCAAUGUCAAGGCCAAUAUCACCACCCCAUUGGCCACCACCACACAGAAAAUGCA ...((((.((((((((((.((((.....)))).))).))))))).))))((....(((((......)))))....(((((((...........))))))).................)). ( -38.30) >DroSec_CAF1 1215 120 - 1 AUUGGCCGCCGAUGGUUGCGGAUCCGGAAUCCUUAGCCCAUCGGAGGUCGCCACCCAUUGUUGCGCCAAUGUCAAGGCCAAUAUCACCACCCCAUUGGCCACCACCACACAGAAAAUGCA ...((((.((((((((((.((((.....)))).))).))))))).))))((....(((((......)))))....(((((((...........))))))).................)). ( -38.30) >DroSim_CAF1 1093 120 - 1 AUUGGCCGCCGAUGGUUGCGGAUCCGGAAUCCUUAGCCCAUCGGAGGUCGCCACCCAUUGUUGCGCCAAUGUCAAGGCCAAUAUCACCACCCCGUUGGCCACCACCACACAGAAAAUGCA ...((((.((((((((((.((((.....)))).))).))))))).))))((....(((((......)))))....(((((((...........))))))).................)). ( -38.60) >DroEre_CAF1 2627 120 - 1 AUUGGCCGCCGAUGGGUCCGGAUCCGGAAUCCUCAGCCCAUCGGAGGUCGCCAGCCAUUGUUGCGCCAAUGUCAAGGCCAAUAUCACCACCCCUUUGGCCUCCACUACACAGAAAAUGCA ...((((.(((((((((..((((.....))))...))))))))).))))((((((....)))).))....((..(((((((.............)))))))..))............... ( -42.82) >DroYak_CAF1 1103 120 - 1 AUUGGCCGCCGAUGGAUCCGGAUCCGGAAUCCUCAGCCCAUCGGAGGUCGCCAGCCAUUGUUGCGCCAAUGUCAAGGCCAAGAUCACCACCCCAUUGGCCACCACCACACACAAAAUGCA ...((((.(((((((.((((....)))).........))))))).))))((..(.(((((......))))).)..((((((.............)))))).................)). ( -37.02) >consensus AUUGGCCGCCGAUGGUUGCGGAUCCGGAAUCCUUAGCCCAUCGGAGGUCGCCACCCAUUGUUGCGCCAAUGUCAAGGCCAAUAUCACCACCCCAUUGGCCACCACCACACAGAAAAUGCA ...((((.(((((((....((((.....)))).....))))))).))))((....(((((......)))))....(((((((...........))))))).................)). (-35.28 = -35.60 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:48 2006