
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,682,087 – 15,682,324 |
| Length | 237 |
| Max. P | 0.999678 |

| Location | 15,682,087 – 15,682,204 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.84 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15682087 117 + 22224390 U-AUAU--GUAAACACAUCGCAUUUGCAUGCAGAUCAGCUUGUAAGCGUAGUUAGAAACUUUGGGAACAAAGUUAGAUACAUAAACUGUCGUUGAGUUUAAUGCGAACGCUGGGAUAAUU .-....--...........((.(((((((.....(((((.................(((((((....))))))).((((.......))))))))).....))))))).)).......... ( -25.70) >DroSec_CAF1 2792 118 + 1 UAAUAU--AUAAACACAUCGCAUUUGCAUGCAGAUCAGCACGUAAGCGUAGUUAGAAACUUUGGGAACAAAGUUAGAUACAUAAACUGUCGUUGAGUUUAAUGUGAACGCUGGGAUAAUU ......--........(((.((.((((.(((......))).))))((((.......(((((((....)))))))...((((((((((.......))))).))))).)))))).))).... ( -27.40) >DroSim_CAF1 2736 118 + 1 UAAUAU--AUAAACACAUCGCAUUUGCAUGCAGAUCAGCUCGUAAGCGUAGUUAGAAACUUUGGGAACAAAGUUAGAUACAUAAACUGUCGUUGAGUUUAAUGUGAACGCUGGGAUAAUU ......--........(((.((.((((..((......))..))))((((.......(((((((....)))))))...((((((((((.......))))).))))).)))))).))).... ( -25.60) >DroEre_CAF1 2771 118 + 1 UAAUAA--AUCAACACAUUGCAUUUGCAUGCAUAUCAGCUUGAAAGCGUAGUUAGAAACUUUGAAAACAAAGUUAGAUACGUAAACUUUCGUUGAGUAUUAUGUGAACGCUGGGAUAAAU ......--(((....((..((.((..((((....(((((..(((((..((((....(((((((....)))))))....)).))..))))))))))....))))..)).)))).))).... ( -24.30) >DroYak_CAF1 2741 120 + 1 UAAUAUAUAUAAACACAUCGCAUUUGCAUGCAGUUCAGCCGGAAAGCGUAGUUAGAAACUUUGGAAACAAAGUUAGAUACAUAAACUUUCGUUGAGUUUUAUGUGAACGCUGGGAUAAAU ...................((.((..((((.(..(((((..(((((..((((....(((((((....)))))))....)).))..))))))))))..).))))..)).)).......... ( -29.40) >consensus UAAUAU__AUAAACACAUCGCAUUUGCAUGCAGAUCAGCUCGUAAGCGUAGUUAGAAACUUUGGGAACAAAGUUAGAUACAUAAACUGUCGUUGAGUUUAAUGUGAACGCUGGGAUAAUU ................(((((((....))))...(((((.................(((((((....)))))))...((((((((((.......))))).)))))...)))))))).... (-22.10 = -22.18 + 0.08)



| Location | 15,682,087 – 15,682,204 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.84 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15682087 117 - 22224390 AAUUAUCCCAGCGUUCGCAUUAAACUCAACGACAGUUUAUGUAUCUAACUUUGUUCCCAAAGUUUCUAACUACGCUUACAAGCUGAUCUGCAUGCAAAUGCGAUGUGUUUAC--AUAU-A ........((((((..((..((((((.......)))))).(((...(((((((....)))))))......)))))..))..))))...(((((....)))))(((((....)--))))-. ( -21.70) >DroSec_CAF1 2792 118 - 1 AAUUAUCCCAGCGUUCACAUUAAACUCAACGACAGUUUAUGUAUCUAACUUUGUUCCCAAAGUUUCUAACUACGCUUACGUGCUGAUCUGCAUGCAAAUGCGAUGUGUUUAU--AUAUUA ...((((.((((((..((((.(((((.......)))))))))....(((((((....))))))).......))))...(((((......)))))....)).)))).......--...... ( -20.80) >DroSim_CAF1 2736 118 - 1 AAUUAUCCCAGCGUUCACAUUAAACUCAACGACAGUUUAUGUAUCUAACUUUGUUCCCAAAGUUUCUAACUACGCUUACGAGCUGAUCUGCAUGCAAAUGCGAUGUGUUUAU--AUAUUA .........(((((..((((.(((((.......)))))))))....(((((((....))))))).......)))))((..(((..(((.((((....)))))))..)))..)--)..... ( -21.20) >DroEre_CAF1 2771 118 - 1 AUUUAUCCCAGCGUUCACAUAAUACUCAACGAAAGUUUACGUAUCUAACUUUGUUUUCAAAGUUUCUAACUACGCUUUCAAGCUGAUAUGCAUGCAAAUGCAAUGUGUUGAU--UUAUUA ...................(((((.(((((((((((.((.((....(((((((....)))))))....)))).)))))).........(((((....)))))....))))).--.))))) ( -23.40) >DroYak_CAF1 2741 120 - 1 AUUUAUCCCAGCGUUCACAUAAAACUCAACGAAAGUUUAUGUAUCUAACUUUGUUUCCAAAGUUUCUAACUACGCUUUCCGGCUGAACUGCAUGCAAAUGCGAUGUGUUUAUAUAUAUUA ........(((((((.......))).....((((((.((.((....(((((((....)))))))....)))).))))))..))))....((((....))))(((((((....))))))). ( -23.50) >consensus AAUUAUCCCAGCGUUCACAUUAAACUCAACGACAGUUUAUGUAUCUAACUUUGUUCCCAAAGUUUCUAACUACGCUUACAAGCUGAUCUGCAUGCAAAUGCGAUGUGUUUAU__AUAUUA ........((((....((((.(((((.......)))))))))....(((((((....))))))).................))))...(((((....))))).................. (-17.72 = -17.80 + 0.08)

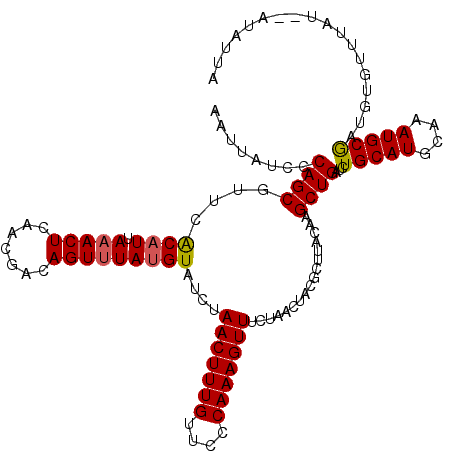
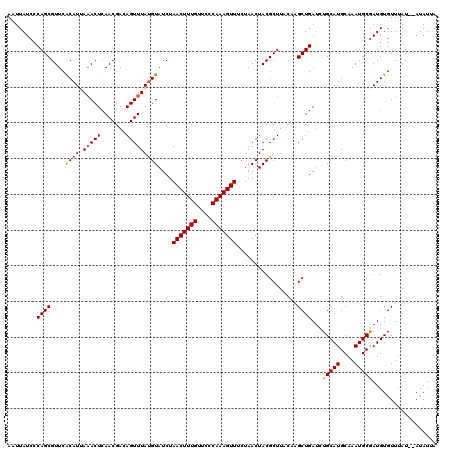
| Location | 15,682,124 – 15,682,244 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -34.18 |
| Energy contribution | -34.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15682124 120 + 22224390 UGUAAGCGUAGUUAGAAACUUUGGGAACAAAGUUAGAUACAUAAACUGUCGUUGAGUUUAAUGCGAACGCUGGGAUAAUUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUU (((.(((((.((....(((((((....)))))))....)).((((((.......))))))......)))))...)))..((((((..(((((((....)))))))...)).))))..... ( -34.10) >DroSec_CAF1 2830 120 + 1 CGUAAGCGUAGUUAGAAACUUUGGGAACAAAGUUAGAUACAUAAACUGUCGUUGAGUUUAAUGUGAACGCUGGGAUAAUUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUU ....(((((.......(((((((....)))))))...((((((((((.......))))).))))).)))))........((((((..(((((((....)))))))...)).))))..... ( -35.80) >DroSim_CAF1 2774 120 + 1 CGUAAGCGUAGUUAGAAACUUUGGGAACAAAGUUAGAUACAUAAACUGUCGUUGAGUUUAAUGUGAACGCUGGGAUAAUUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUU ....(((((.......(((((((....)))))))...((((((((((.......))))).))))).)))))........((((((..(((((((....)))))))...)).))))..... ( -35.80) >DroEre_CAF1 2809 120 + 1 UGAAAGCGUAGUUAGAAACUUUGAAAACAAAGUUAGAUACGUAAACUUUCGUUGAGUAUUAUGUGAACGCUGGGAUAAAUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUU ....(((((.......(((((((....)))))))...(((((((((((.....)))).))))))).))))).....(((((((((..(((((((....)))))))...)).))))))).. ( -37.00) >DroYak_CAF1 2781 120 + 1 GGAAAGCGUAGUUAGAAACUUUGGAAACAAAGUUAGAUACAUAAACUUUCGUUGAGUUUUAUGUGAACGCUGGGAUAAAUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUU ....(((((.......(((((((....)))))))...(((((((((((.....))).)))))))).))))).....(((((((((..(((((((....)))))))...)).))))))).. ( -41.70) >consensus CGUAAGCGUAGUUAGAAACUUUGGGAACAAAGUUAGAUACAUAAACUGUCGUUGAGUUUAAUGUGAACGCUGGGAUAAUUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUU ....(((((.......(((((((....)))))))...((((((((((.......))))).))))).)))))........((((((..(((((((....)))))))...)).))))..... (-34.18 = -34.42 + 0.24)

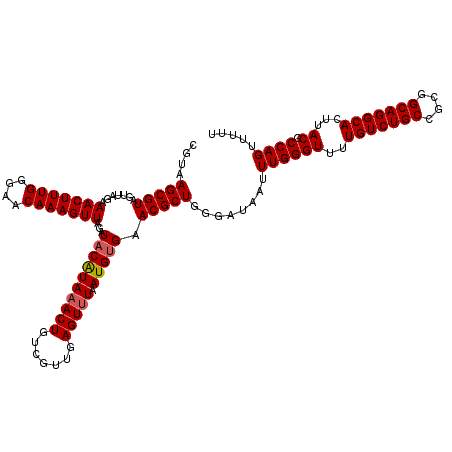
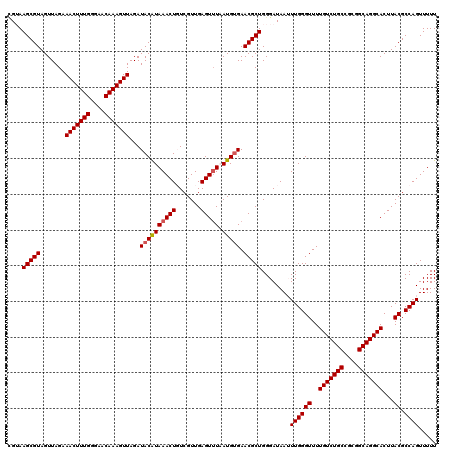
| Location | 15,682,124 – 15,682,244 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -23.66 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15682124 120 - 22224390 AAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCAAAUUAUCCCAGCGUUCGCAUUAAACUCAACGACAGUUUAUGUAUCUAACUUUGUUCCCAAAGUUUCUAACUACGCUUACA ......(((.((...((.((((....)))).))..))))).........(((((..((((.(((((.......)))))))))....(((((((....))))))).......))))).... ( -25.00) >DroSec_CAF1 2830 120 - 1 AAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCAAAUUAUCCCAGCGUUCACAUUAAACUCAACGACAGUUUAUGUAUCUAACUUUGUUCCCAAAGUUUCUAACUACGCUUACG ......(((.((...((.((((....)))).))..))))).........(((((..((((.(((((.......)))))))))....(((((((....))))))).......))))).... ( -24.70) >DroSim_CAF1 2774 120 - 1 AAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCAAAUUAUCCCAGCGUUCACAUUAAACUCAACGACAGUUUAUGUAUCUAACUUUGUUCCCAAAGUUUCUAACUACGCUUACG ......(((.((...((.((((....)))).))..))))).........(((((..((((.(((((.......)))))))))....(((((((....))))))).......))))).... ( -24.70) >DroEre_CAF1 2809 120 - 1 AAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCAAUUUAUCCCAGCGUUCACAUAAUACUCAACGAAAGUUUACGUAUCUAACUUUGUUUUCAAAGUUUCUAACUACGCUUUCA ..(((.(((.((...((.((((....)))).))..))))).))).....(((((.......((((..(((....)))...))))..(((((((....))))))).......))))).... ( -25.90) >DroYak_CAF1 2781 120 - 1 AAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCAAUUUAUCCCAGCGUUCACAUAAAACUCAACGAAAGUUUAUGUAUCUAACUUUGUUUCCAAAGUUUCUAACUACGCUUUCC ..(((.(((.((...((.((((....)))).))..))))).))).....(((((..((((((......((....))))))))....(((((((....))))))).......))))).... ( -25.70) >consensus AAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCAAAUUAUCCCAGCGUUCACAUUAAACUCAACGACAGUUUAUGUAUCUAACUUUGUUCCCAAAGUUUCUAACUACGCUUACA ......(((.((...((.((((....)))).))..))))).........(((((..((((.(((((.......)))))))))....(((((((....))))))).......))))).... (-23.66 = -23.90 + 0.24)

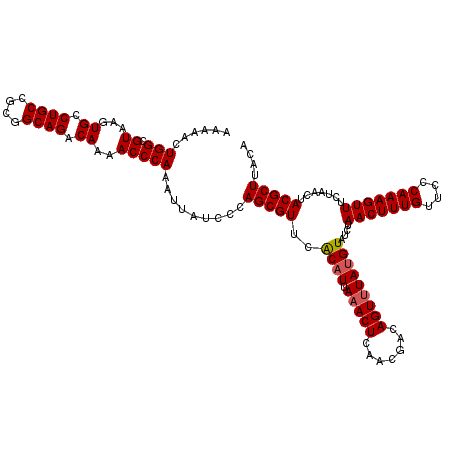

| Location | 15,682,164 – 15,682,284 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -29.88 |
| Energy contribution | -30.76 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15682164 120 + 22224390 AUAAACUGUCGUUGAGUUUAAUGCGAACGCUGGGAUAAUUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUAAUUUGACGAUGGAACACCUAAGCCCUUUCAU .....(((((((..((((((((..(((.(((((((....))..((..(((((((....)))))))...)).))))).)))..))))..))))..)))))))................... ( -32.10) >DroSec_CAF1 2870 120 + 1 AUAAACUGUCGUUGAGUUUAAUGUGAACGCUGGGAUAAUUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUAAUUUGACGGCGGAACACUUAAGCCCUUUCAU .....(((((((..((((((((.((((.(((((((....))..((..(((((((....)))))))...)).))))).)))).))))..))))..)))))))................... ( -35.00) >DroSim_CAF1 2814 120 + 1 AUAAACUGUCGUUGAGUUUAAUGUGAACGCUGGGAUAAUUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUAAUUUGACGGCGGAACACUUAAGCCCUUUCAU .....(((((((..((((((((.((((.(((((((....))..((..(((((((....)))))))...)).))))).)))).))))..))))..)))))))................... ( -35.00) >DroEre_CAF1 2849 120 + 1 GUAAACUUUCGUUGAGUAUUAUGUGAACGCUGGGAUAAAUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUUAAUUGACGGCAGAACACUUAAGUCCUUUCAU ((((((((.....)))).))))(((((....(((((...((((((....((((((......(((......))).(((.((((.....))))..)))))))))..)))))))))))))))) ( -27.90) >DroYak_CAF1 2821 120 + 1 AUAAACUUUCGUUGAGUUUUAUGUGAACGCUGGGAUAAAUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUUAAUUGACGGCAGAACACUUAAGCCCUUUCAU ..((((((.....))))))...(((((....(((.....((((((....((((((......(((......))).(((.((((.....))))..)))))))))..)))))).))).))))) ( -29.00) >consensus AUAAACUGUCGUUGAGUUUAAUGUGAACGCUGGGAUAAUUUGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUAAUUUGACGGCGGAACACUUAAGCCCUUUCAU .....(((((((..((((((((.((((.(((((((....))..((..(((((((....)))))))...)).))))).)))).))))..))))..)))))))................... (-29.88 = -30.76 + 0.88)

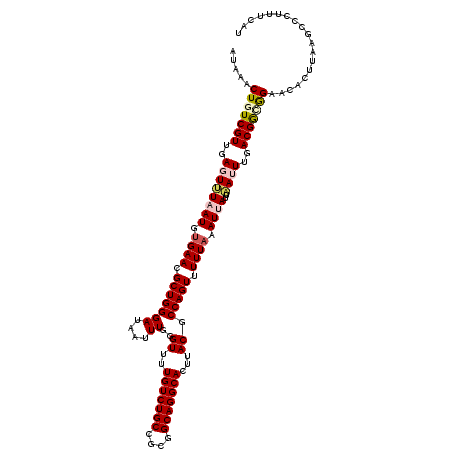
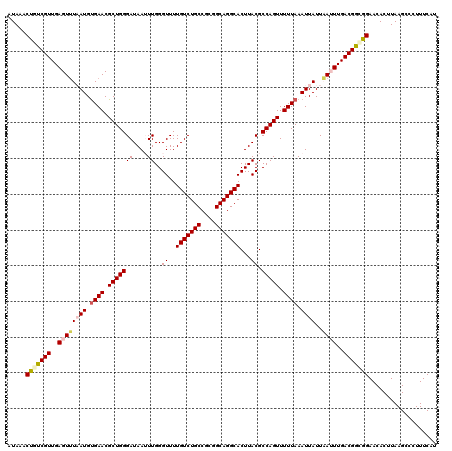
| Location | 15,682,204 – 15,682,324 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -24.92 |
| Energy contribution | -24.96 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15682204 120 + 22224390 UGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUAAUUUGACGAUGGAACACCUAAGCCCUUUCAUCUGAAAAAUAUAUUCAAAUAUGACGCAAUAUGAAACAUAA .(((((((((((((....)))))))......(((....((((((.....))))))...))).......))))))((((((.((....((((......))))....))..))))))..... ( -25.50) >DroSec_CAF1 2910 120 + 1 UGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUAAUUUGACGGCGGAACACUUAAGCCCUUUCAUCUGAAAAAUAUAUUCAAAUAUGACGCAAUAUGAAACAUAA .(((((((((((((....)))))))....((((.(((.((((....))))...)))))))........))))))((((((.((....((((......))))....))..))))))..... ( -30.00) >DroSim_CAF1 2854 120 + 1 UGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUAAUUUGACGGCGGAACACUUAAGCCCUUUCAUCUGAAAAAUAUAUUCAAAUAUGACGCAAUAUGAAACAUAA .(((((((((((((....)))))))....((((.(((.((((....))))...)))))))........))))))((((((.((....((((......))))....))..))))))..... ( -30.00) >DroEre_CAF1 2889 120 + 1 UGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUUAAUUGACGGCAGAACACUUAAGUCCUUUCAUCUGAAAAAUAUGUUCAAAUAUGACGCAAUAUGAAACACAA .((..(((((((((....))))))).....(((.(((.((((.....))))..)))))).........))..))((((((.((....((((......))))....))..))))))..... ( -24.40) >DroYak_CAF1 2861 120 + 1 UGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUUAAUUGACGGCAGAACACUUAAGCCCUUUCAUCUGAAAAAUAUAUUCAAAUAUGACGCAAUAUGAAACAAAA .(((((((((((((....))))))).....(((.(((.((((.....))))..)))))).........))))))((((((.((....((((......))))....))..))))))..... ( -27.80) >consensus UGGGUUUUGUCUGCCGCGGCAGGCACUUACGCCAGUUUUUAAAUUAUUAAUUUGACGGCGGAACACUUAAGCCCUUUCAUCUGAAAAAUAUAUUCAAAUAUGACGCAAUAUGAAACAUAA .(((((((((((((((.....(((......))).....((((((.....)))))))))))).)))...))))))((((((.((....((((......))))....))..))))))..... (-24.92 = -24.96 + 0.04)

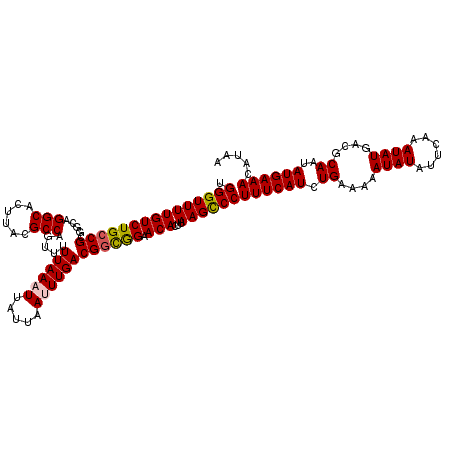
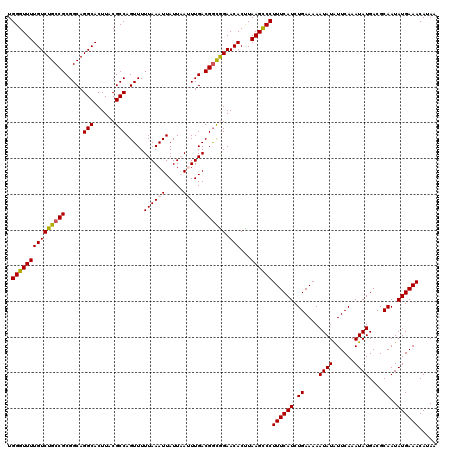
| Location | 15,682,204 – 15,682,324 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -24.68 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15682204 120 - 22224390 UUAUGUUUCAUAUUGCGUCAUAUUUGAAUAUAUUUUUCAGAUGAAAGGGCUUAGGUGUUCCAUCGUCAAAUUAAUAAUUUAAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCA ....((((....((((((((..(((((((.((((..(..(((((..((((......))))..)))))..)..)))))))))))...)))))))).((.((((....)))).))))))... ( -27.90) >DroSec_CAF1 2910 120 - 1 UUAUGUUUCAUAUUGCGUCAUAUUUGAAUAUAUUUUUCAGAUGAAAGGGCUUAAGUGUUCCGCCGUCAAAUUAAUAAUUUAAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCA ....((((....((((((((..(((((((.((((..(..((((...((((......))))...))))..)..)))))))))))...)))))))).((.((((....)))).))))))... ( -25.40) >DroSim_CAF1 2854 120 - 1 UUAUGUUUCAUAUUGCGUCAUAUUUGAAUAUAUUUUUCAGAUGAAAGGGCUUAAGUGUUCCGCCGUCAAAUUAAUAAUUUAAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCA ....((((....((((((((..(((((((.((((..(..((((...((((......))))...))))..)..)))))))))))...)))))))).((.((((....)))).))))))... ( -25.40) >DroEre_CAF1 2889 120 - 1 UUGUGUUUCAUAUUGCGUCAUAUUUGAACAUAUUUUUCAGAUGAAAGGACUUAAGUGUUCUGCCGUCAAUUAAAUAAUUUAAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCA ((((....(((.((((((((..((((((..(((((....((((..(((((......)))))..))))....))))).))))))...))))))))))).((((....))))))))...... ( -29.20) >DroYak_CAF1 2861 120 - 1 UUUUGUUUCAUAUUGCGUCAUAUUUGAAUAUAUUUUUCAGAUGAAAGGGCUUAAGUGUUCUGCCGUCAAUUAAAUAAUUUAAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCA (((((((.....((((((((..(((((((.(((((....((((..(((((......)))))..))))....))))))))))))...)))))))).((((......))))))))))).... ( -31.40) >consensus UUAUGUUUCAUAUUGCGUCAUAUUUGAAUAUAUUUUUCAGAUGAAAGGGCUUAAGUGUUCCGCCGUCAAAUUAAUAAUUUAAAAACUGGCGUAAGUGCCUGCCGCGGCAGACAAAACCCA ....((((....((((((((..(((((((.((((.....((((...((((......))))...)))).....)))))))))))...)))))))).((.((((....)))).))))))... (-24.68 = -24.72 + 0.04)

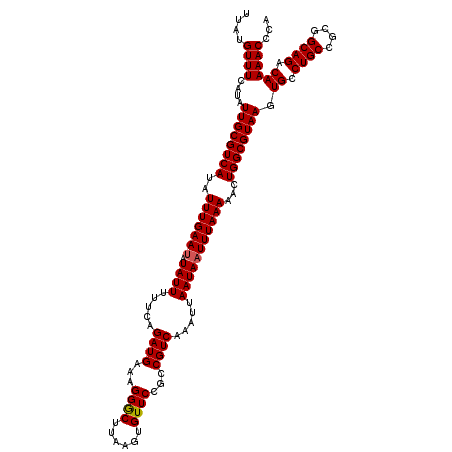

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:46 2006