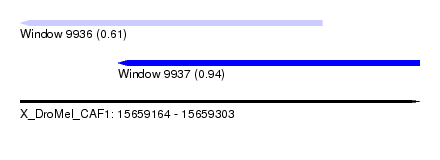
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,659,164 – 15,659,303 |
| Length | 139 |
| Max. P | 0.943029 |

| Location | 15,659,164 – 15,659,269 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -18.39 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
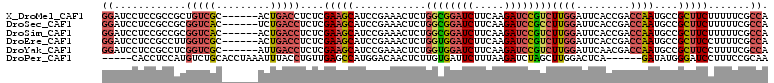
>X_DroMel_CAF1 15659164 105 - 22224390 GGAUCCUCCGCCGCUGUCGC------ACUGACCUCUCGAAGCAUCCGAAACUCUGGCGGAUCUUCAAGAUCCGUCUUGGAUUCACCGACCAAUGCCGCUUCUUUUUCGCCA ((.....))((.((.(((((------..(((....)))..))....(((.((..((((((((.....))))))))..)).)))...)))....)).))............. ( -27.80) >DroSec_CAF1 1916 105 - 1 GGAUCCUCCGCCGCGGUCAC------UCUGACCUCUCGAAGCAUCCGAAACUCUGGCGGAUCUUCAAGAUCCGCCUUGGAUUCACCGACCAAUGCCGCUUCUUUUUCGCCA ((.....))...((((((..------...))))....(((((....(((.((..((((((((.....))))))))..)).)))...(........))))))......)).. ( -34.40) >DroSim_CAF1 912 105 - 1 GGAUCCUCCGCCGCGGUCAC------ACUGACCUCUCGAAGCAUCCGAAACUCUGGCGGAUCUUCAAGAUCCGUCUUGGAUUCACCGACCAAUGCCGCUUCUUUUUCGCCA ((.....))...((((((..------...))))....(((((....(((.((..((((((((.....))))))))..)).)))...(........))))))......)).. ( -31.70) >DroEre_CAF1 1841 105 - 1 GGAUCCUCCGCCUUGGUCGC------ACUGACCUCUCGAAGCAUCCGAAACUCUGGUGGAUCUUCAAGAUCCGUCUUGGAUUCACCGACCAAUGCCGCUUCCUUUUCGCCA (((....(.((.((((((((------..(((....)))..))............((((((((..(((((....))))))))))))))))))).)).)..)))......... ( -31.30) >DroYak_CAF1 1037 105 - 1 GGAUCCUCCGCCUCGGUCGC------AUUGACCUCUCGAAGCAUCCGAAACUCUGGUGGAUCUUCAAGAUCCGUCUUGGAUUCAACGACCAAUGCCGCUUCCUUUUCGCCA ((((((......((((..((------.((((....)))).))..))))......(..(((((.....)))))..)..))))))............................ ( -27.30) >DroPer_CAF1 1620 100 - 1 -----CACCUCCAUGUCUGCACCUAAAUUUACCUGUUGAGCCAUGGACAACUCUUGUGAUUCUUUAAGAUCUAGCUUGGACUCA------GAUAUGGGAUCCUUUCCGCAA -----....((((((((((..((..(((......)))((((..((((....(((((........)))))))))))))))...))------))))))))............. ( -19.90) >consensus GGAUCCUCCGCCGCGGUCGC______ACUGACCUCUCGAAGCAUCCGAAACUCUGGCGGAUCUUCAAGAUCCGUCUUGGAUUCACCGACCAAUGCCGCUUCCUUUUCGCCA ((............(((((.........)))))....(((((............((((((((.....))))))))((((.........))))....))))).......)). (-18.39 = -19.28 + 0.89)


| Location | 15,659,198 – 15,659,303 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -21.24 |
| Energy contribution | -22.63 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
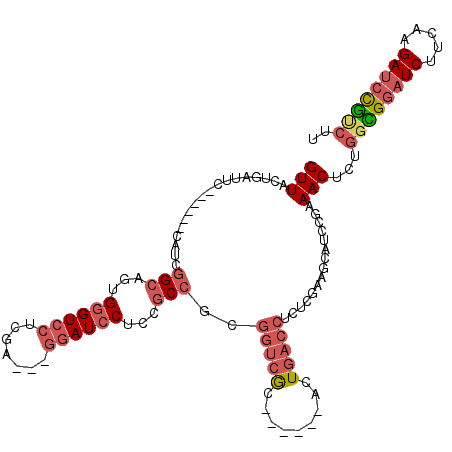
>X_DroMel_CAF1 15659198 105 - 22224390 GUUACUGAUUC------CAUCGGCGUUGGGUCCUCGUCGUGGAUCCUCCGCCGCUGUCGC------ACUGACCUCUCGAAGCAUCCGAAACUCUGGCGGAUCUUCAAGAUCCGUCUU ((((.((....------((.(((((..((((((.......))))))..))))).))...)------).))))...(((.......)))......((((((((.....)))))))).. ( -38.20) >DroSec_CAF1 1950 105 - 1 GUUACUGAUUC------CAUUGGCGUUGGGUCCUCGACGUGGAUCCUCCGCCGCGGUCAC------UCUGACCUCUCGAAGCAUCCGAAACUCUGGCGGAUCUUCAAGAUCCGCCUU (((.(.(((.(------....((((..((((((.......))))))..))))..((((..------...)))).......).))).).)))...((((((((.....)))))))).. ( -41.50) >DroSim_CAF1 946 105 - 1 GUUACUGAUUC------CAUCGGCGUUGGGUCCUCGACGUGGAUCCUCCGCCGCGGUCAC------ACUGACCUCUCGAAGCAUCCGAAACUCUGGCGGAUCUUCAAGAUCCGUCUU (((.(.(((.(------...(((((..((((((.......))))))..))))).((((..------...)))).......).))).).)))...((((((((.....)))))))).. ( -40.00) >DroEre_CAF1 1875 102 - 1 GUUACUGAUUC------CAUCGGCAGUGGGUCCUCGA---GGAUCCUCCGCCUUGGUCGC------ACUGACCUCUCGAAGCAUCCGAAACUCUGGUGGAUCUUCAAGAUCCGUCUU ((((.((...(------((..(((.(.((((((....---)))))).).))).)))...)------).)))).(((.((((.(((((.........))))))))).)))........ ( -32.70) >DroYak_CAF1 1071 102 - 1 GUUACUGAUUC------CACCGGCAGUGGGUCCUCGA---GGAUCCUCCGCCUCGGUCGC------AUUGACCUCUCGAAGCAUCCGAAACUCUGGUGGAUCUUCAAGAUCCGUCUU .....(((.((------(((((((.(.((((((....---)))))).).)))((((..((------.((((....)))).))..))))......))))))...)))........... ( -39.20) >DroPer_CAF1 1648 105 - 1 GUUGCUGACAAUUUUGGCACCAGCAGCGA--CCAC----------CACCUCCAUGUCUGCACCUAAAUUUACCUGUUGAGCCAUGGACAACUCUUGUGAUUCUUUAAGAUCUAGCUU (((((((.............)))))))..--....----------....((((((.((.((.(...........).)))).))))))....(((((........)))))........ ( -19.12) >consensus GUUACUGAUUC______CAUCGGCAGUGGGUCCUCGA___GGAUCCUCCGCCGCGGUCGC______ACUGACCUCUCGAAGCAUCCGAAACUCUGGCGGAUCUUCAAGAUCCGUCUU (((..................(((...((((((.......))))))...)))..(((((.........)))))...............)))...((((((((.....)))))))).. (-21.24 = -22.63 + 1.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:33 2006