
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,657,945 – 15,658,050 |
| Length | 105 |
| Max. P | 0.664504 |

| Location | 15,657,945 – 15,658,050 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 67.96 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.10 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15657945 105 + 22224390 G---AUGAUUUAAAUGGCGACUCCU--UUCUUCCCCAGAUACGAAAGCAACGAGAAUGUCGUGCUCGUGUGCUCCACAAUCGUUUGCUCCUUUGGCAAGCAAGUGGUGGA .---..........(((.((.....--....)).))).((((((..(((.(((.....))))))))))))..(((((....(((((((.....))))))).....))))) ( -25.50) >DroVir_CAF1 638 80 + 1 -----------------------------C-CUUGCAGAUAUGAAAGUAAUGAGAACAUUGAGCUCGUCUGCUCGACAAUUGUCUGCUCGUUUGGCAAACAGGUUGUGGA -----------------------------.-...((((((..(....(((((....)))))..)..))))))((.((((((((.((((.....)))).))).))))).)) ( -21.50) >DroEre_CAF1 685 98 + 1 ----------UUAAUAAUAACGCAU--UUCAUCCGCAGAUACGAAAGCAACGAGAAUGUCGUGCUCGUGUGCUCCACAAUCGUUUGCUCUUUUGGCAAGCAGGUGGUGGA ----------...............--.......(((..(((((..(((.(((.....))))))))))))))(((((.((((((((((.....))))))).))).))))) ( -27.20) >DroWil_CAF1 592 107 + 1 UGCUCUUAUUUCAAAUUCAAUUUC---UCAUCUCAUAGAUAUGAAAGCAAUGAGAAUAUUGAACUUGUCUGCUCGACAAUCGUUUGCUCCUUUGGCAAACAGGUGGUGGA .((........(((.(((((((((---((((.((((....)))).....))))))).)))))).)))...))((.((.((((((((((.....))))))).))).)).)) ( -24.40) >DroMoj_CAF1 673 91 + 1 ------------------AAUUCCUCACUC-CACACAGAUAUGAAAGCAAUGAGAACAUUGAGCUCGUCUGCUCGACAAUCGUCUGCUCGUUCGGCAAACAGGUGGUGGA ------------------...(((.....(-(((.......((((((((((((.....((((((......))))))...)))).))))..))))........)))).))) ( -22.86) >DroAna_CAF1 333 92 + 1 ----------UCAUUAUUAAAUCGU--U------GCAGAUAUGAGAGUAAUGAGAACGUAGUACUCGUUUGCUCAACGAUCGUUUGCUCAUUCGGUAAACAGGUAGUGGA ----------((((((((..(((((--(------(.((..(((((.(..(((....)))..).)))))...))))))))).(((((((.....))))))).)))))))). ( -24.80) >consensus __________U_AAU___AACUCCU__UUC_CCCGCAGAUAUGAAAGCAAUGAGAACAUUGAGCUCGUCUGCUCGACAAUCGUUUGCUCCUUUGGCAAACAGGUGGUGGA ................................((((.(((.....(((((((((.........))))).)))).....)))(((((((.....))))))).....)))). (-16.70 = -16.10 + -0.60)

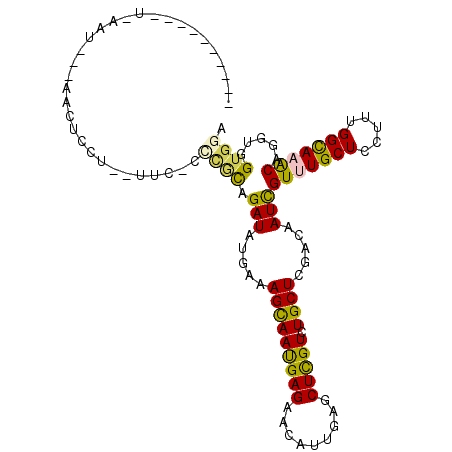

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:30 2006