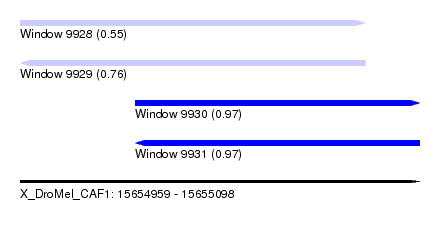
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,654,959 – 15,655,098 |
| Length | 139 |
| Max. P | 0.974127 |

| Location | 15,654,959 – 15,655,079 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -35.49 |
| Consensus MFE | -22.14 |
| Energy contribution | -21.84 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
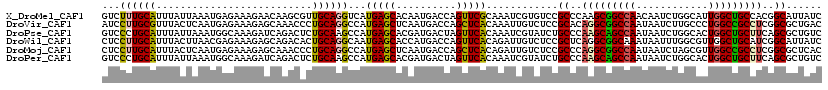
>X_DroMel_CAF1 15654959 120 + 22224390 GUCUUUGCAUUUAUUAAAUGAGAAAGAACAAGCGUUGCAGGUCAUGAGCACAAUGACCAGUUCGCAAAUCGUGUCCGCCCAAGCGGCCAACAAUCUGGCAUUGGCUGCCACGGCAUUAUC .(((((.((((.....))))..)))))....(((..((.((((((.......)))))).)).)))...........(((...((((((((..........))))))))...)))...... ( -34.60) >DroVir_CAF1 67223 120 + 1 AUCCUUGCGUUUACUCAAUGAGAAAGAGCAAACCCUGCAGGCCAUGAGCUCAAUGACCAGCUCACAAAUUGUCUCCGCACAGGCGGCCAAUAAUCUUGCCCUGGCCGCCUCGGCGCUGAC ...(((((((((.(((.........))).))))...)))))...((((((........))))))......(((..(((..(((((((((............)))))))))..)))..))) ( -40.30) >DroPse_CAF1 49285 120 + 1 GUCCCUGCAUUUAUUAAAUGGCAAAGAUCAGACUCUGCAAGCCAUGAGCACGAUGACUAGUUCACAAAUCGUAUCUGCCCAAGCAGCCAAUAAUCUGGCACUGGCUGCUUCAGCGCUGUC .................(((((..(((......)))....))))).((((((((.............)))))....((..(((((((((............)))))))))..)))))... ( -30.32) >DroWil_CAF1 7884 120 + 1 CUCCUUGCAUUUACUUAACGAGAAAGAGCAGACACUGCAGGCAAUGAGCACCAUGACCAGUUCACAGAUUGUCUCCGCUCAGGCGGCAAAUAAUUUGGCGUUGGCUGCAUCGGCAUUAUC (((.(((........))).))).....((.((...(((((.((((((((..........)))).((((((((..((((....))))...))))))))..)))).))))))).))...... ( -30.90) >DroMoj_CAF1 71480 120 + 1 CUCCUUGCAUUUACUCAAUGAGAAAGAGCAAACCCUGCAGGCCAUGAGCUCAAUGACCAGCUCACAGAUUGUCUCCGCCCAGGCGGCCAAUAAUCUAGCGUUGGCCGCCUCGGCGCUCAC (((.(((........))).)))...((((.......((((....((((((........))))))....))))....(((.(((((((((((........))))))))))).))))))).. ( -46.50) >DroPer_CAF1 60354 120 + 1 GUCCCUGCAUUUAUUAAAUGGCAAAGAUCAGACUCUGCAAGCCAUGAGCACGAUGACUAGUUCACAAAUCGUAUCUGCCCAAGCAGCCAAUAAUCUGGCACUGGCUGCUUCAGCGCUGUC .................(((((..(((......)))....))))).((((((((.............)))))....((..(((((((((............)))))))))..)))))... ( -30.32) >consensus GUCCUUGCAUUUACUAAAUGAGAAAGAGCAAACCCUGCAGGCCAUGAGCACAAUGACCAGUUCACAAAUCGUCUCCGCCCAAGCGGCCAAUAAUCUGGCACUGGCUGCCUCGGCGCUGUC ...((((((..........................))))))...(((((..........)))))............((..(((((((((............)))))))))..))...... (-22.14 = -21.84 + -0.30)
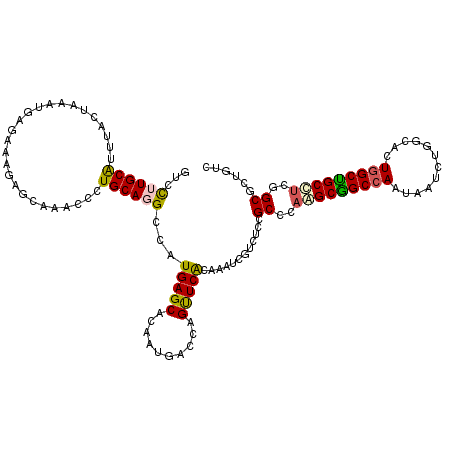

| Location | 15,654,959 – 15,655,079 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -30.19 |
| Energy contribution | -29.37 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15654959 120 - 22224390 GAUAAUGCCGUGGCAGCCAAUGCCAGAUUGUUGGCCGCUUGGGCGGACACGAUUUGCGAACUGGUCAUUGUGCUCAUGACCUGCAACGCUUGUUCUUUCUCAUUUAAUAAAUGCAAAGAC ......(((..(((.((((((........)))))).)))..)))(((((.(..(((((....((((((.......)))))))))))..).)))))......................... ( -32.90) >DroVir_CAF1 67223 120 - 1 GUCAGCGCCGAGGCGGCCAGGGCAAGAUUAUUGGCCGCCUGUGCGGAGACAAUUUGUGAGCUGGUCAUUGAGCUCAUGGCCUGCAGGGUUUGCUCUUUCUCAUUGAGUAAACGCAAGGAU (((..(((..((((((((((..........))))))))))..)))..)))...(..((((((.(....).))))))..)(((...(.((((((((.........)))))))).).))).. ( -51.80) >DroPse_CAF1 49285 120 - 1 GACAGCGCUGAAGCAGCCAGUGCCAGAUUAUUGGCUGCUUGGGCAGAUACGAUUUGUGAACUAGUCAUCGUGCUCAUGGCUUGCAGAGUCUGAUCUUUGCCAUUUAAUAAAUGCAGGGAC ..((((.(((((((((((((((......))))))))))))((((((((..((((.(....).))))))).)))))........))).).))).(((((((.(((.....)))))))))). ( -41.00) >DroWil_CAF1 7884 120 - 1 GAUAAUGCCGAUGCAGCCAACGCCAAAUUAUUUGCCGCCUGAGCGGAGACAAUCUGUGAACUGGUCAUGGUGCUCAUUGCCUGCAGUGUCUGCUCUUUCUCGUUAAGUAAAUGCAAGGAG .......((..((((((....))....(((((((.((...(((((((.((...((((((.....))))))(((.........))))).))))))).....)).))))))).)))).)).. ( -29.90) >DroMoj_CAF1 71480 120 - 1 GUGAGCGCCGAGGCGGCCAACGCUAGAUUAUUGGCCGCCUGGGCGGAGACAAUCUGUGAGCUGGUCAUUGAGCUCAUGGCCUGCAGGGUUUGCUCUUUCUCAUUGAGUAAAUGCAAGGAG .....((((.((((((((((..........)))))))))).))))........(((((((((.(....).)))))))))(((...(.((((((((.........)))))))).).))).. ( -53.30) >DroPer_CAF1 60354 120 - 1 GACAGCGCUGAAGCAGCCAGUGCCAGAUUAUUGGCUGCUUGGGCAGAUACGAUUUGUGAACUAGUCAUCGUGCUCAUGGCUUGCAGAGUCUGAUCUUUGCCAUUUAAUAAAUGCAGGGAC ..((((.(((((((((((((((......))))))))))))((((((((..((((.(....).))))))).)))))........))).).))).(((((((.(((.....)))))))))). ( -41.00) >consensus GACAGCGCCGAGGCAGCCAAUGCCAGAUUAUUGGCCGCCUGGGCGGAGACAAUUUGUGAACUGGUCAUUGUGCUCAUGGCCUGCAGAGUCUGCUCUUUCUCAUUUAAUAAAUGCAAGGAC ......(((.((((((((((..........)))))))))).)))((((....(((((.....((((((.(....))))))).))))).....))))........................ (-30.19 = -29.37 + -0.83)



| Location | 15,654,999 – 15,655,098 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -20.52 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15654999 99 + 22224390 GUCAUGAGCACAAUGACCAGUUCGCAAAUCGUGUCCGCCCAAGCGGCCAACAAUCUGGCAUUGGCUGCCACGGCAUUAUCGGCAGCAGGUGGUGGUGGU .((((.(.(((((((.((((..(((.....))).((((....))))........)))))))).((((((...........))))))..))).).)))). ( -32.30) >DroPse_CAF1 49325 84 + 1 GCCAUGAGCACGAUGACUAGUUCACAAAUCGUAUCUGCCCAAGCAGCCAAUAAUCUGGCACUGGCUGCUUCAGCGCUGUCCGUG--------------- ..((((((((((((.............)))))....((..(((((((((............)))))))))..)))))...))))--------------- ( -24.02) >DroGri_CAF1 62224 84 + 1 GCCAUGAGCUCAAUGACCAGCUCACAAAUUGUCUCCGCACAGGCGGCCAAUAAUCUUGCCCUGGCCGCCUCGGCGCUGACGGCG--------------- (((.((((((........))))))......(((..(((..(((((((((............)))))))))..)))..)))))).--------------- ( -39.60) >DroWil_CAF1 7924 87 + 1 GCAAUGAGCACCAUGACCAGUUCACAGAUUGUCUCCGCUCAGGCGGCAAAUAAUUUGGCGUUGGCUGCAUCGGCAUUAUCAGCGGUG------------ ........((((.(((((((..(.((((((((..((((....))))...)))))))))..)))).(((....)))...)))..))))------------ ( -24.70) >DroMoj_CAF1 71520 84 + 1 GCCAUGAGCUCAAUGACCAGCUCACAGAUUGUCUCCGCCCAGGCGGCCAAUAAUCUAGCGUUGGCCGCCUCGGCGCUCACGGCG--------------- (((.(((((.....(((...(.....)...)))...(((.(((((((((((........))))))))))).)))))))).))).--------------- ( -43.10) >DroPer_CAF1 60394 84 + 1 GCCAUGAGCACGAUGACUAGUUCACAAAUCGUAUCUGCCCAAGCAGCCAAUAAUCUGGCACUGGCUGCUUCAGCGCUGUCCGUG--------------- ..((((((((((((.............)))))....((..(((((((((............)))))))))..)))))...))))--------------- ( -24.02) >consensus GCCAUGAGCACAAUGACCAGUUCACAAAUCGUCUCCGCCCAAGCGGCCAAUAAUCUGGCACUGGCUGCCUCGGCGCUGUCGGCG_______________ ((..(((((..........)))))............((..(((((((((............)))))))))..)).......))................ (-20.52 = -19.80 + -0.72)



| Location | 15,654,999 – 15,655,098 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -20.94 |
| Energy contribution | -20.28 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15654999 99 - 22224390 ACCACCACCACCUGCUGCCGAUAAUGCCGUGGCAGCCAAUGCCAGAUUGUUGGCCGCUUGGGCGGACACGAUUUGCGAACUGGUCAUUGUGCUCAUGAC .............(((((((.........)))))))(((((((((..((((.(((.....))).))))((.....))..)))).))))).......... ( -30.80) >DroPse_CAF1 49325 84 - 1 ---------------CACGGACAGCGCUGAAGCAGCCAGUGCCAGAUUAUUGGCUGCUUGGGCAGAUACGAUUUGUGAACUAGUCAUCGUGCUCAUGGC ---------------(((((.....(((.((((((((((((......)))))))))))).)))......((((.(....).)))).)))))........ ( -32.80) >DroGri_CAF1 62224 84 - 1 ---------------CGCCGUCAGCGCCGAGGCGGCCAGGGCAAGAUUAUUGGCCGCCUGUGCGGAGACAAUUUGUGAGCUGGUCAUUGAGCUCAUGGC ---------------.((((((..(((..((((((((((..........))))))))))..)))..))).....(((((((.(....).)))))))))) ( -41.60) >DroWil_CAF1 7924 87 - 1 ------------CACCGCUGAUAAUGCCGAUGCAGCCAACGCCAAAUUAUUUGCCGCCUGAGCGGAGACAAUCUGUGAACUGGUCAUGGUGCUCAUUGC ------------....((.......))(((((.(((....((((.........((((....)))).(((.....(....)..))).)))))))))))). ( -19.30) >DroMoj_CAF1 71520 84 - 1 ---------------CGCCGUGAGCGCCGAGGCGGCCAACGCUAGAUUAUUGGCCGCCUGGGCGGAGACAAUCUGUGAGCUGGUCAUUGAGCUCAUGGC ---------------.((((((((((((.((((((((((..........)))))))))).)))(....)..((.((((.....)))).))))))))))) ( -48.40) >DroPer_CAF1 60394 84 - 1 ---------------CACGGACAGCGCUGAAGCAGCCAGUGCCAGAUUAUUGGCUGCUUGGGCAGAUACGAUUUGUGAACUAGUCAUCGUGCUCAUGGC ---------------(((((.....(((.((((((((((((......)))))))))))).)))......((((.(....).)))).)))))........ ( -32.80) >consensus _______________CGCCGACAGCGCCGAGGCAGCCAAUGCCAGAUUAUUGGCCGCCUGGGCGGAGACAAUUUGUGAACUGGUCAUUGUGCUCAUGGC ...............(((.......(((.((((((((((..........)))))))))).)))(((.....)))))).....(((((.(....)))))) (-20.94 = -20.28 + -0.66)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:27 2006