
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,653,517 – 15,653,625 |
| Length | 108 |
| Max. P | 0.669104 |

| Location | 15,653,517 – 15,653,625 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.92 |
| Mean single sequence MFE | -38.99 |
| Consensus MFE | -31.31 |
| Energy contribution | -32.37 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15653517 108 + 22224390 GCAUAAUGU------------UUUCAGUUGGACCCGGCACCAUACCAUCACCGUGGACACCAGUGAAUGCCGGUCCUCCAGGCGCACUUGGAUCGGCAGACACAAAUGGCAGCAUGGUGG .(((.((((------------(....((((....)))).((((.((((....))))......(((..(((((((((...((.....)).)))))))))..)))..)))).))))).))). ( -36.00) >DroVir_CAF1 57226 114 + 1 UCUCUGCUA------UUUGUUUUGCAGUCGGACCCGGCACCAUACCAUCCCCGUGGCCACCAGUGAAUGCCGGUCCUCCUGGUCCCCUGGGAUCGGCAGACACAAAUGGCAGCAUGGUGG ...((((((------((((((((((((..(((((.((((.....((((....)))).((....))..)))))))))..))(((((....))))).))))).)))))))))))........ ( -45.90) >DroGri_CAF1 52837 120 + 1 UUUCUGCUUGGUUUUUUUCCAUUACAGUCGGACCCGGCACCAUACCAUCUCCGUGGCCACCAGUGAAUGCCGGUCCUCCUGGUCCCCUGGGAUCGGCAGACACAAAUGGCAGCAUGGUGG ....((((.(((((...............))))).)))).....(((((...((.((((...(((..((((((((((...((...)).))))))))))..)))...)))).))..))))) ( -41.96) >DroWil_CAF1 2 90 + 1 ------------------------------GACCCGGCACCAUACCAUCUCCGUGGACACCAGUGAAUGCCGGUCCUCCGGGUCCUCUGGGAUCGGCAGACACAAAUGGCAGCAUGGUGG ------------------------------.......((((((.((((....))))...((((((..((((((((((..(....)...))))))))))..)))...)))....)))))). ( -33.30) >DroMoj_CAF1 59587 108 + 1 UCAUUGCUA------------UUGCAGUUGGACCCGGCACCAUACCAUCUCCGUGGCCACCAGUGAAUGCCGGUCCUCCUGGUCCCCUGGGAUCGGCAGACACAAAUGGCAGCAUGGUGG ....(((..------------..)))((((....))))......(((((...((.((((...(((..((((((((((...((...)).))))))))))..)))...)))).))..))))) ( -40.90) >DroPer_CAF1 44334 102 + 1 CCAU------------------UACAGUCGGACCCGGCACCAUACCAUCUCCGUGGACCCCAGUGAAUGCCGGUCCUCCAGGUGCACUAGGUUCGGCAGACACAAAUGGAAGUAUGGUGG ((..------------------....((((....))))(((((((....(((((((...)).(((..(((((..(((..((.....)))))..)))))..)))..))))).))))))))) ( -35.90) >consensus UCAUUGCU_____________UUACAGUCGGACCCGGCACCAUACCAUCUCCGUGGACACCAGUGAAUGCCGGUCCUCCUGGUCCCCUGGGAUCGGCAGACACAAAUGGCAGCAUGGUGG ..........................((((....))))....((((((((((((((...)).(((..((((((((((...(.....).))))))))))..)))..)))).)).)))))). (-31.31 = -32.37 + 1.06)

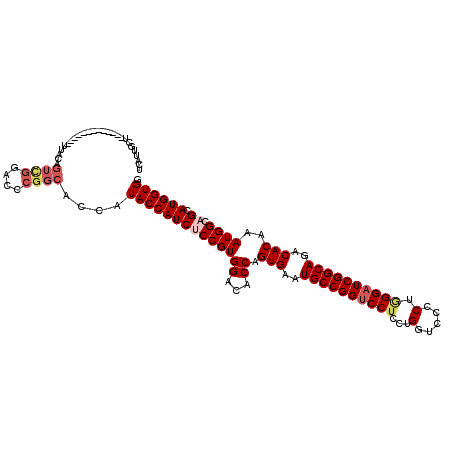

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:24 2006