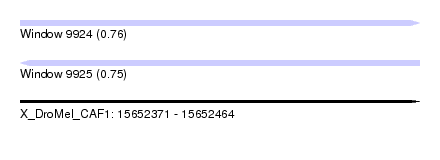
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,652,371 – 15,652,464 |
| Length | 93 |
| Max. P | 0.764605 |

| Location | 15,652,371 – 15,652,464 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 75.44 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -18.59 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
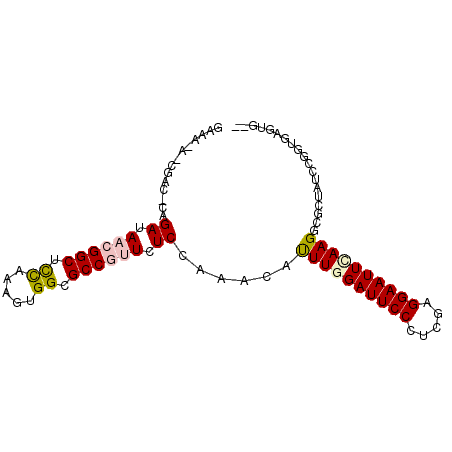
>X_DroMel_CAF1 15652371 93 + 22224390 GGAAUAUCACCAGCGAUAUCGGCUUUACAGUGGCGCCCUUCUCAAAUCAUUUAGAUUCCCUCGAGGAAUUCAAGGAGUUAUCGGGUGAGUGGA ......(((((..(((((.((((...........))).........((.(((..(((((.....)))))..)))))).))))))))))..... ( -19.80) >DroVir_CAF1 55060 92 + 1 GAAA-AGCGACUUAGACAACGGCUCCAAAGUGGCGCCGUUCUCCAAACAUUUGGAUUCCCUCGAGGAAUUCAAGGCUCUCACCGGUGAGUUGU ....-.((((((((((.((((((.((.....)).)))))).))......((((((((((.....))))))))))...........)))))))) ( -31.00) >DroGri_CAF1 51115 90 + 1 GAAA-AACGACACAGACAACGGCUCCAAAGUGGCGCCGUUCUCCAAACAUUUGGAUUCCCUCGAGGAAUUCAAGGCUCUAUCCGGUGAGUU-- ....-.........((.((((((.((.....)).)))))).))......((((((((((.....))))))))))((((........)))).-- ( -26.90) >DroSim_CAF1 3 81 + 1 ------------GCGAUAUCGGCUUUACAGUGGCGCCCUUCUCAAAUCAUUUAGAUUCCCUCGAGGAAUUCAAGGAGUUAUCCGGUGAGUGGA ------------(((.(((.(......).))).)))((..((((......((..(((((.....)))))..))(((....)))..)))).)). ( -17.40) >DroMoj_CAF1 56841 90 + 1 GAAG-AGCGACUCAGAUAACGGCUCCAAAUUGGCGCCGUUCUCCAAGCACUUGGAUUCCCUCGAGGAAUUUAAGACCCUGUCCGGUGAGUG-- ....-....((((.((.((((((.((.....)).)))))).))......((((((((((.....))))))))))(((......))))))).-- ( -29.50) >DroPer_CAF1 43933 84 + 1 AAAUU-------AAGAAAACGGCUUUAAGUUGGCGCCGUUCUCUAACCACUUGGAUUCCCUUGAGGAAUUCAAGGCAUUAUCUGGUGAGUU-- .....-------.(((.((((((...........)))))).))).((((((((((((((.....))))))))))........)))).....-- ( -24.20) >consensus GAAA_A_CGAC_CAGAUAACGGCUCCAAAGUGGCGCCGUUCUCCAAACAUUUGGAUUCCCUCGAGGAAUUCAAGGCGCUAUCCGGUGAGUG__ ..............((.((((((.((.....)).)))))).))......((((((((((.....))))))))))................... (-18.59 = -18.78 + 0.20)

| Location | 15,652,371 – 15,652,464 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 75.44 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -15.25 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
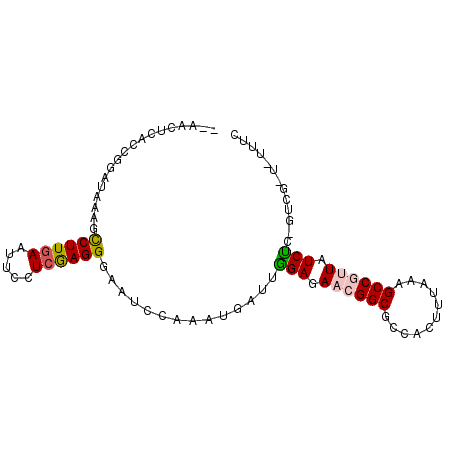
>X_DroMel_CAF1 15652371 93 - 22224390 UCCACUCACCCGAUAACUCCUUGAAUUCCUCGAGGGAAUCUAAAUGAUUUGAGAAGGGCGCCACUGUAAAGCCGAUAUCGCUGGUGAUAUUCC .....((((((((((...((((((.....))))))(((((.....)))))......(((...........)))..)))))..)))))...... ( -23.70) >DroVir_CAF1 55060 92 - 1 ACAACUCACCGGUGAGAGCCUUGAAUUCCUCGAGGGAAUCCAAAUGUUUGGAGAACGGCGCCACUUUGGAGCCGUUGUCUAAGUCGCU-UUUC ....((((....))))(((.(((.(((((.....))))).)))..(((((((.((((((.((.....)).)))))).)))))).))))-.... ( -31.90) >DroGri_CAF1 51115 90 - 1 --AACUCACCGGAUAGAGCCUUGAAUUCCUCGAGGGAAUCCAAAUGUUUGGAGAACGGCGCCACUUUGGAGCCGUUGUCUGUGUCGUU-UUUC --........((((....((((((.....))))))..))))(((((..((((.((((((.((.....)).)))))).))))...))))-)... ( -29.80) >DroSim_CAF1 3 81 - 1 UCCACUCACCGGAUAACUCCUUGAAUUCCUCGAGGGAAUCUAAAUGAUUUGAGAAGGGCGCCACUGUAAAGCCGAUAUCGC------------ ....(((...........((((((.....))))))(((((.....))))))))...(((...........)))........------------ ( -16.70) >DroMoj_CAF1 56841 90 - 1 --CACUCACCGGACAGGGUCUUAAAUUCCUCGAGGGAAUCCAAGUGCUUGGAGAACGGCGCCAAUUUGGAGCCGUUAUCUGAGUCGCU-CUUC --...((.((....((((........))))...)))).....((((((..((.((((((.((.....)).)))))).))..)).))))-.... ( -29.30) >DroPer_CAF1 43933 84 - 1 --AACUCACCAGAUAAUGCCUUGAAUUCCUCAAGGGAAUCCAAGUGGUUAGAGAACGGCGCCAACUUAAAGCCGUUUUCUU-------AAUUU --(((.(((..(((..(.((((((.....)))))).))))...))))))((((((((((...........)))))))))).-------..... ( -25.20) >consensus __AACUCACCGGAUAAAGCCUUGAAUUCCUCGAGGGAAUCCAAAUGAUUGGAGAACGGCGCCACUUUAAAGCCGUUAUCUC_GUCG_U_UUUC ..................((((((.....))))))..............(((.((((((...........)))))).)))............. (-15.25 = -15.28 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:22 2006