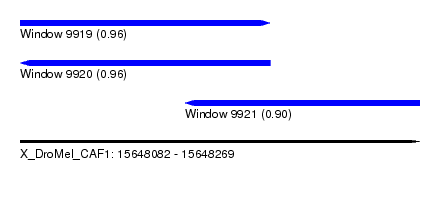
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,648,082 – 15,648,269 |
| Length | 187 |
| Max. P | 0.964882 |

| Location | 15,648,082 – 15,648,199 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.64 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -23.65 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
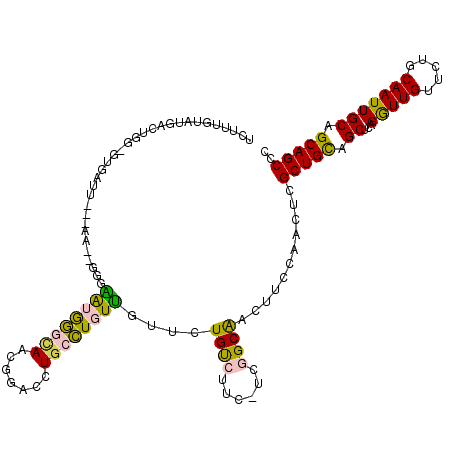
>X_DroMel_CAF1 15648082 117 + 22224390 AGCGAAACCUAAAGGAAAUCUACGAAAAAAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCGAGCACUUGCAGCACUGGGCUGCUGCAAUUGCAGAACAACCUGAGCUGCAGCGAGU .......((....)).................(((.(((((.(((......)))..(((((..(((.((((((((......)))))))).)))((......)))))))))))).))) ( -37.10) >DroVir_CAF1 42335 92 + 1 -----------------------AA-U-AAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCAAGCACUUGCGGCACGGGGCUGCUGCAAUUGCAGAACAAUCUCAGCAACAGCGAAU -----------------------..-.-...((((((.((.(((.....)))))))))))...(((.((((((((......)))))))).)))............((....)).... ( -27.80) >DroPse_CAF1 27608 96 + 1 ---GAACCC------------------CAUAAGCUGGCUGCAUGAAAAACAUAACCAGCUCGAGCACCUGCAGCACGGGGCUGCUGCAAUUGCAGAACAAUCUAAACAACAGGGAAU ---....((------------------(...((((((.(..(((.....))).)))))))...(((..(((((((......)))))))..)))..................)))... ( -28.30) >DroGri_CAF1 38763 91 + 1 -----------------------A--U-AUAAACUGGCUGCAUGAAAAACAUCACCAGCUCAAGCACUUGCGGCACGGGGCUGCUGCAAUUGCAGAACAAUCUCAGCAACAGCGAAU -----------------------.--.-.....((((((((((((......)))...((....))...)))))).))).((((.(((...((.(((....)))))))).)))).... ( -23.90) >DroMoj_CAF1 43379 103 + 1 ------------AGG-UACUUACAG-UAAAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCAAGCACUUGCGGCACGGGGCUGCUGCAAUUGCAGAACAAUCUCAGCAACAGCGAAU ------------(((-(........-.....((((((.((.(((.....)))))))))))...(((.((((((((......)))))))).)))......))))..((....)).... ( -28.30) >DroPer_CAF1 26960 96 + 1 ---GAACCC------------------CAUAAGCUGGCUGCAUGAAAAACAUAACCAGCUCGAGCACCUGCAGCACGGGGCUGCUGCAAUUGCAGAACAAUCUAAACAACAGGGAAU ---....((------------------(...((((((.(..(((.....))).)))))))...(((..(((((((......)))))))..)))..................)))... ( -28.30) >consensus _______________________A__U_AAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCAAGCACUUGCAGCACGGGGCUGCUGCAAUUGCAGAACAAUCUCAGCAACAGCGAAU ...............................((((((.((.(((.....)))))))))))...(((.((((((((......)))))))).)))........................ (-23.65 = -24.23 + 0.58)



| Location | 15,648,082 – 15,648,199 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.64 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -27.67 |
| Energy contribution | -27.53 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15648082 117 - 22224390 ACUCGCUGCAGCUCAGGUUGUUCUGCAAUUGCAGCAGCCCAGUGCUGCAAGUGCUCGAGCUGGUGAUGUUUUUCAUGCAGCCAGCUUUUUUUUCGUAGAUUUCCUUUAGGUUUCGCU ....((((((((.(.(((((..((((....)))))))))..).))))).)))((..((((((((.(((.....)))...))))))))..........((...((....))..)))). ( -38.50) >DroVir_CAF1 42335 92 - 1 AUUCGCUGUUGCUGAGAUUGUUCUGCAAUUGCAGCAGCCCCGUGCCGCAAGUGCUUGAGCUGGUGAUGUUUUUCAUGCAGCCAGCUUUU-A-UU----------------------- ...((..(((((((..((((.....))))..)))))))..)).(((....).))..((((((((.(((.....)))...))))))))..-.-..----------------------- ( -32.10) >DroPse_CAF1 27608 96 - 1 AUUCCCUGUUGUUUAGAUUGUUCUGCAAUUGCAGCAGCCCCGUGCUGCAGGUGCUCGAGCUGGUUAUGUUUUUCAUGCAGCCAGCUUAUG------------------GGGUUC--- ...((((((.....(((....)))(((.((((((((......)))))))).)))..((((((((((((.....))))..)))))))))))------------------)))...--- ( -34.90) >DroGri_CAF1 38763 91 - 1 AUUCGCUGUUGCUGAGAUUGUUCUGCAAUUGCAGCAGCCCCGUGCCGCAAGUGCUUGAGCUGGUGAUGUUUUUCAUGCAGCCAGUUUAU-A--U----------------------- ...((..(((((((..((((.....))))..)))))))..)).(((....).)).(((((((((.(((.....)))...))))))))).-.--.----------------------- ( -31.20) >DroMoj_CAF1 43379 103 - 1 AUUCGCUGUUGCUGAGAUUGUUCUGCAAUUGCAGCAGCCCCGUGCCGCAAGUGCUUGAGCUGGUGAUGUUUUUCAUGCAGCCAGCUUUUUA-CUGUAAGUA-CCU------------ ....((((((((...(((((.....)))))))))))))...((((....((((...((((((((.(((.....)))...))))))))..))-))....)))-)..------------ ( -35.70) >DroPer_CAF1 26960 96 - 1 AUUCCCUGUUGUUUAGAUUGUUCUGCAAUUGCAGCAGCCCCGUGCUGCAGGUGCUCGAGCUGGUUAUGUUUUUCAUGCAGCCAGCUUAUG------------------GGGUUC--- ...((((((.....(((....)))(((.((((((((......)))))))).)))..((((((((((((.....))))..)))))))))))------------------)))...--- ( -34.90) >consensus AUUCGCUGUUGCUGAGAUUGUUCUGCAAUUGCAGCAGCCCCGUGCCGCAAGUGCUCGAGCUGGUGAUGUUUUUCAUGCAGCCAGCUUAU_A__U_______________________ ....((((((((...(((((.....)))))))))))))...(..(.....)..)..((((((((.(((.....)))...)))))))).............................. (-27.67 = -27.53 + -0.14)
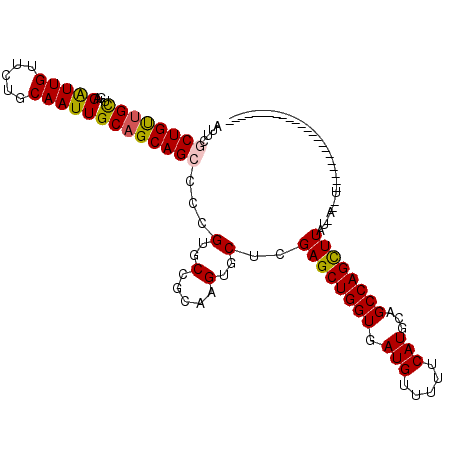
| Location | 15,648,159 – 15,648,269 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.99 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -19.17 |
| Energy contribution | -17.90 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15648159 110 - 22224390 UCUUUGUAUGACUGG-GUAAUUC--------GGAUGGGCAACGGACCUGCCUGUUGUUCUGUCUUC-UCGGCAACUUCCAACUCGCUGCAGCUCAGGUUGUUCUGCAAUUGCAGCAGCCC .............((-((.....--------(((.(((((((((......)))))))))((((...-..))))...))).....(((((((..((((....))))...))))))).)))) ( -35.10) >DroVir_CAF1 42387 92 - 1 -------------------------GA--GGGUGUAAAAAACGCACCUGCUUCUGGUU-UGCCAACAACAGCGACGUCCAAUUCGCUGUUGCUGAGAUUGUUCUGCAAUUGCAGCAGCCC -------------------------..--((((((.......))))))(((.(((..(-(((((.(((((((((........))))))))).))(((....)))))))...))).))).. ( -32.00) >DroSec_CAF1 4887 110 - 1 UCUUUGUAUGACUGG-GUGAUUU--------GGAUGGGCAACGGACCUGCCUGUUGUUCUGUCUUC-UCGGCAACUUCCAACUCGCUGCAGCUCAGGUUGUUCUGCAAUUGCAGCAGCCC ........(((((((-((((.((--------(((.(((((((((......)))))))))((((...-..))))...))))).))))).))).)))(((((..((((....))))))))). ( -39.80) >DroEre_CAF1 4507 116 - 1 UCUUUGUAUGACUGG-GUGAUUGGAAA--GGGAAUGGGCAACGGACCUGCCUGUUGUUCUGUCUUC-UCGGCAACUUCCAACUCGCUGCAGCUCAGGUUGUUCUGCAAUUGCAGCAGCCC ........(((((((-((((((((((.--(((((.(((((((((......)))))))))....)))-))(....))))))).))))).))).)))(((((..((((....))))))))). ( -42.60) >DroYak_CAF1 5154 119 - 1 UCUUUGUAUGAUUCGAAUGAGGGGGAAUGGGGAAUGGGCAACGGACCUGCCUGUUGUUCUGUCUUC-UCGGCAACUUCCAACUCGCUGCAGCUCAGGUUGUUCUGCAAUUGCAGCAGCCC ..................(((..((((.(((((..(((((((((......)))))))))..)))))-..(....)))))..)))(((((((..((((....))))...)))))))..... ( -39.90) >DroMoj_CAF1 43442 92 - 1 -------------------------AA--UGGUGUAAAUAACGCACCUGCUUCUGGUU-UGCCAACAACAGCGACGUCCAAUUCGCUGUUGCUGAGAUUGUUCUGCAAUUGCAGCAGCCC -------------------------..--.(((((.......))))).(((.(((..(-(((((.(((((((((........))))))))).))(((....)))))))...))).))).. ( -30.60) >consensus UCUUUGUAUGACUGG_GUGAUU___AA__GGGAAUGGGCAACGGACCUGCCUGUUGUUCUGUCUUC_UCGGCAACUUCCAACUCGCUGCAGCUCAGGUUGUUCUGCAAUUGCAGCAGCCC ................................((((((((.......))))))))....((((......))))...........(((((.((...(((((.....))))))).))))).. (-19.17 = -17.90 + -1.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:18 2006