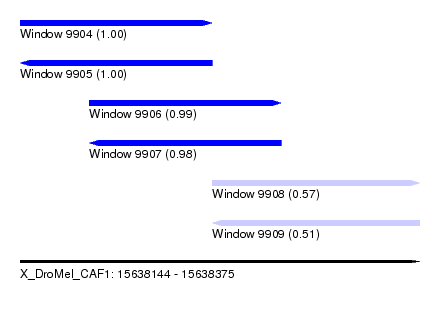
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,638,144 – 15,638,375 |
| Length | 231 |
| Max. P | 0.999741 |

| Location | 15,638,144 – 15,638,255 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.07 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -14.36 |
| Energy contribution | -16.16 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15638144 111 + 22224390 AAAUGAAAGAGAAAAACGGAUUCUCUUUAUGUAAAAUCUUAGAAUUAACGCUCUCUUAGGUUCCACA---------AUUGGUUCCGAUUGGAACCGCGCACAAAGAGAGCGAGGAAGAGA .....((((((((.......))))))))....................(((((((((.(((((((..---------((((....)))))))))))(....).)))))))))......... ( -32.90) >DroSec_CAF1 2839 90 + 1 AACUGAAAGAGAA---------------------AAUCUUAGAAUUAACGCUCUCUUGGGUUCCACA---------AUUGCUUCCGAUUGGAACAGCGCACAAAGAGAGCGACGAAGAGA ......((((...---------------------..))))........(((((((((..((((((..---------((((....))))))))))........)))))))))......... ( -22.80) >DroSim_CAF1 2835 90 + 1 AACUGAAAGAGAA---------------------AAUCUUAGAAUGAACGCUCUCUUGGGUUCCACA---------AUUGCUUCCGAUUGGAACCGCGCACAAAGAGAGCGACGAAGAGA ......((((...---------------------..))))........(((((((((.(((((((..---------((((....))))))))))).......)))))))))......... ( -25.20) >DroEre_CAF1 2500 114 + 1 AAAUGAAAGAGA---ACAAAUUCUCUUUAUGCAAAAACU-AAAAUUUACGCUCUCUUAGGUUCCACAAGUGUUCCAAUUGGUUCCAAUUGGAACCGCGCACGAAGAGAGA--GAAAGAGA .....(((((((---(....)))))))).........((-....(((...(((((((.(....)....((((((((((((....))))))))).))).....))))))).--.))).)). ( -31.40) >DroYak_CAF1 2163 95 + 1 AAAUGGAAGAGAA--ACCCAUUCUCUUUAUGUAAAAACUGAAAAUUUCCACUCUCUUAGGU---------------------UCCAAUUGGAACCGCGCACGAAGAGAGG--CAAAGAGA ...((((((((((--.....)))))((((.((....))))))...)))))(((((((.(((---------------------(((....)))))).......))))))).--........ ( -26.10) >consensus AAAUGAAAGAGAA__AC__AUUCUCUUUAUG_AAAAUCUUAGAAUUAACGCUCUCUUAGGUUCCACA_________AUUGCUUCCGAUUGGAACCGCGCACAAAGAGAGCGACGAAGAGA ................................................(((((((((.(((((((.......................))))))).......)))))))))......... (-14.36 = -16.16 + 1.80)


| Location | 15,638,144 – 15,638,255 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.07 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -13.12 |
| Energy contribution | -14.92 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.47 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15638144 111 - 22224390 UCUCUUCCUCGCUCUCUUUGUGCGCGGUUCCAAUCGGAACCAAU---------UGUGGAACCUAAGAGAGCGUUAAUUCUAAGAUUUUACAUAAAGAGAAUCCGUUUUUCUCUUUCAUUU .........(((((((((.((.(((((((((....))))))...---------.)))..))..)))))))))....................((((((((.......))))))))..... ( -32.00) >DroSec_CAF1 2839 90 - 1 UCUCUUCGUCGCUCUCUUUGUGCGCUGUUCCAAUCGGAAGCAAU---------UGUGGAACCCAAGAGAGCGUUAAUUCUAAGAUU---------------------UUCUCUUUCAGUU .........(((((((((.(......((((((..(....)....---------..))))))).)))))))))..((((..((((..---------------------...))))..)))) ( -20.40) >DroSim_CAF1 2835 90 - 1 UCUCUUCGUCGCUCUCUUUGUGCGCGGUUCCAAUCGGAAGCAAU---------UGUGGAACCCAAGAGAGCGUUCAUUCUAAGAUU---------------------UUCUCUUUCAGUU ..((((.(.(((((((((.....(.(((((((..(....)....---------..)))))))))))))))))......).))))..---------------------............. ( -23.90) >DroEre_CAF1 2500 114 - 1 UCUCUUUC--UCUCUCUUCGUGCGCGGUUCCAAUUGGAACCAAUUGGAACACUUGUGGAACCUAAGAGAGCGUAAAUUUU-AGUUUUUGCAUAAAGAGAAUUUGU---UCUCUUUCAUUU ........--.(((((((..(.(((.((((((((((....))))))))))....))).)....))))))).(((((....-....)))))..((((((((....)---)))))))..... ( -34.40) >DroYak_CAF1 2163 95 - 1 UCUCUUUG--CCUCUCUUCGUGCGCGGUUCCAAUUGGA---------------------ACCUAAGAGAGUGGAAAUUUUCAGUUUUUACAUAAAGAGAAUGGGU--UUCUCUUCCAUUU ((((((((--..((((((.(....)((((((....)))---------------------))).))))))((((((((.....)))))))).))))))))(((((.--......))))).. ( -29.90) >consensus UCUCUUCGUCGCUCUCUUUGUGCGCGGUUCCAAUCGGAACCAAU_________UGUGGAACCUAAGAGAGCGUUAAUUCUAAGAUUUU_CAUAAAGAGAAU__GU__UUCUCUUUCAUUU .........(((((((((.(....)(((((((.......................))))))).)))))))))................................................ (-13.12 = -14.92 + 1.80)


| Location | 15,638,184 – 15,638,295 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -21.94 |
| Energy contribution | -23.94 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15638184 111 + 22224390 AGAAUUAACGCUCUCUUAGGUUCCACA---------AUUGGUUCCGAUUGGAACCGCGCACAAAGAGAGCGAGGAAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUGGCCAGAAAAAU ........(((((((((.(((((((..---------((((....))))))))))).(((.(.....).)))...)))))))))......((((..((((........))))..))))... ( -35.50) >DroSec_CAF1 2858 111 + 1 AGAAUUAACGCUCUCUUGGGUUCCACA---------AUUGCUUCCGAUUGGAACAGCGCACAAAGAGAGCGACGAAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUGGCCAGAAAAAU ........(((((((((.(((((((..---------((((....))))))))))..(((.(.....).))).).)))))))))......((((..((((........))))..))))... ( -32.00) >DroSim_CAF1 2854 111 + 1 AGAAUGAACGCUCUCUUGGGUUCCACA---------AUUGCUUCCGAUUGGAACCGCGCACAAAGAGAGCGACGAAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUGGCCAGAAAAAU ........(((((((((.(((((((..---------((((....)))))))))))((((.(.....).))).).)))))))))......((((..((((........))))..))))... ( -33.40) >DroEre_CAF1 2536 107 + 1 AAAAUUUACGCUCUCUUAGGUUCCACAAGUGUUCCAAUUGGUUCCAAUUGGAACCGCGCACGAAGAGAGA--GAAAGAGAGCGA--GAGUUUCGAGUCGAAUUGGAAUGGC--------- ........(((((((((...(((.....((((((((((((....)))))))).....)))).....))).--..))))))))).--..(((((((......)))))))...--------- ( -34.60) >DroYak_CAF1 2201 86 + 1 AAAAUUUCCACUCUCUUAGGU---------------------UCCAAUUGGAACCGCGCACGAAGAGAGG--CAAAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUG----------- ....(((((.(((((((.(((---------------------(((....))))))..((.(.......))--).))))))).).))))(((((((......))))))).----------- ( -28.10) >consensus AGAAUUAACGCUCUCUUAGGUUCCACA_________AUUGCUUCCGAUUGGAACCGCGCACAAAGAGAGCGACGAAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUGGCCAGAAAAAU ........(((((((((.(((((((.......................))))))).(((.(.....).)))...))))))))).....(((((((......)))))))............ (-21.94 = -23.94 + 2.00)

| Location | 15,638,184 – 15,638,295 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -18.16 |
| Energy contribution | -19.80 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15638184 111 - 22224390 AUUUUUCUGGCCAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUUCCUCGCUCUCUUUGUGCGCGGUUCCAAUCGGAACCAAU---------UGUGGAACCUAAGAGAGCGUUAAUUCU .......(((.....))).((((...)))).......(((((((((((.(((.(.....).))).((((((....))))))...---------...))).....))))))))........ ( -29.20) >DroSec_CAF1 2858 111 - 1 AUUUUUCUGGCCAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUUCGUCGCUCUCUUUGUGCGCUGUUCCAAUCGGAAGCAAU---------UGUGGAACCCAAGAGAGCGUUAAUUCU .......(((.....))).((((...)))).......(((((((((.(.(((.(.....).)))).((((((..(....)....---------..))))))..)))))))))........ ( -26.10) >DroSim_CAF1 2854 111 - 1 AUUUUUCUGGCCAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUUCGUCGCUCUCUUUGUGCGCGGUUCCAAUCGGAAGCAAU---------UGUGGAACCCAAGAGAGCGUUCAUUCU .......(((.....))).((((...)))).......(((((((((.(.(((.(.....).))))(((((((..(....)....---------..))))))).)))))))))........ ( -30.20) >DroEre_CAF1 2536 107 - 1 ---------GCCAUUCCAAUUCGACUCGAAACUC--UCGCUCUCUUUC--UCUCUCUUCGUGCGCGGUUCCAAUUGGAACCAAUUGGAACACUUGUGGAACCUAAGAGAGCGUAAAUUUU ---------..........((((...))))....--.(((((((((..--..(((...((....))((((((((((....))))))))))......)))....)))))))))........ ( -31.10) >DroYak_CAF1 2201 86 - 1 -----------CAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUUUG--CCUCUCUUCGUGCGCGGUUCCAAUUGGA---------------------ACCUAAGAGAGUGGAAAUUUU -----------..((((..((((...))))........((((((((.(--((.(.....).).))((((((....)))---------------------))).))))))))))))..... ( -25.60) >consensus AUUUUUCUGGCCAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUUCGUCGCUCUCUUUGUGCGCGGUUCCAAUCGGAACCAAU_________UGUGGAACCUAAGAGAGCGUUAAUUCU ...................((((...)))).......(((((((((...(((.(.....).))).(((((((.......................))))))).)))))))))........ (-18.16 = -19.80 + 1.64)


| Location | 15,638,255 – 15,638,375 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -29.57 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15638255 120 + 22224390 GCGAGAGAGUUUCGAGUCGAAUUGGAAUGGCCAGAAAAAUAUAUUUCUGCUUUCAGUUUCGAAAAAUUUUUCGAUGGAAGCAGCCGAGUGCGAAUGCGAAUGCUCAAUGAGAAGAGCUUU ......((((((((..(((..((((.......(((((......)))))(((((((...((((((....)))))))))))))..))))...)))...)))).))))............... ( -31.50) >DroSec_CAF1 2929 120 + 1 GCGAGAGAGUUUCGAGUCGAAUUGGAAUGGCCAGAAAAAUAUAUUUCUGCUUUCAGUUUCGAAAAAUUUUUGGAUGGAAGCAGCCGAGUGCGAAUGCGAAUGCUCANNNNNNNNNNNNNN ......((((((((..(((..((((.......(((((......)))))(((((((..((((((.....)))))))))))))..))))...)))...)))).))))............... ( -29.30) >DroSim_CAF1 2925 120 + 1 GCGAGAGAGUUUCGAGUCGAAUUGGAAUGGCCAGAAAAAUAUAUUUCUGCUUUCAGUUUCGAAAAAUUUUUCGAUGGAAGCAGCCGAGUGCGAAUGCGAAUGCUCAAUGAGAAGAGCUUU ......((((((((..(((..((((.......(((((......)))))(((((((...((((((....)))))))))))))..))))...)))...)))).))))............... ( -31.50) >consensus GCGAGAGAGUUUCGAGUCGAAUUGGAAUGGCCAGAAAAAUAUAUUUCUGCUUUCAGUUUCGAAAAAUUUUUCGAUGGAAGCAGCCGAGUGCGAAUGCGAAUGCUCAAUGAGAAGAGCUUU ......((((((((..(((..((((.......(((((......)))))(((((((...((((((....)))))))))))))..))))...)))...)))).))))............... (-29.57 = -29.90 + 0.33)


| Location | 15,638,255 – 15,638,375 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -20.73 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15638255 120 - 22224390 AAAGCUCUUCUCAUUGAGCAUUCGCAUUCGCACUCGGCUGCUUCCAUCGAAAAAUUUUUCGAAACUGAAAGCAGAAAUAUAUUUUUCUGGCCAUUCCAAUUCGACUCGAAACUCUCUCGC ...((((........)))).(((((....))....((((((((.((((((((....))))))...)).))))(((((......)))))))))...............))).......... ( -26.50) >DroSec_CAF1 2929 120 - 1 NNNNNNNNNNNNNNUGAGCAUUCGCAUUCGCACUCGGCUGCUUCCAUCCAAAAAUUUUUCGAAACUGAAAGCAGAAAUAUAUUUUUCUGGCCAUUCCAAUUCGACUCGAAACUCUCUCGC ...............(((..((((((..((....))..))).................(((((...(((.(((((((......))))).))..)))...)))))...))).)))...... ( -17.40) >DroSim_CAF1 2925 120 - 1 AAAGCUCUUCUCAUUGAGCAUUCGCAUUCGCACUCGGCUGCUUCCAUCGAAAAAUUUUUCGAAACUGAAAGCAGAAAUAUAUUUUUCUGGCCAUUCCAAUUCGACUCGAAACUCUCUCGC ...((((........)))).(((((....))....((((((((.((((((((....))))))...)).))))(((((......)))))))))...............))).......... ( -26.50) >consensus AAAGCUCUUCUCAUUGAGCAUUCGCAUUCGCACUCGGCUGCUUCCAUCGAAAAAUUUUUCGAAACUGAAAGCAGAAAUAUAUUUUUCUGGCCAUUCCAAUUCGACUCGAAACUCUCUCGC .............((((((....((....))....((((((((.((((((((....))))))...)).))))(((((......)))))))))..........).)))))........... (-20.73 = -21.40 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:08 2006