
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,637,262 – 15,637,458 |
| Length | 196 |
| Max. P | 0.896671 |

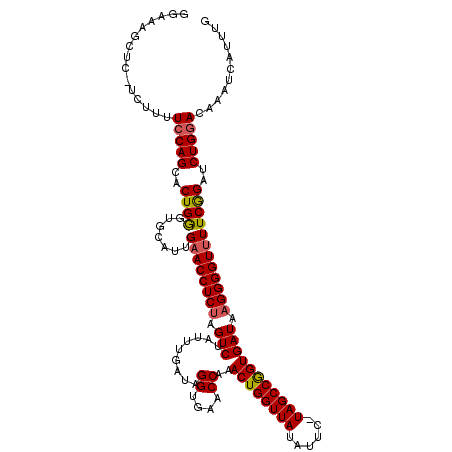
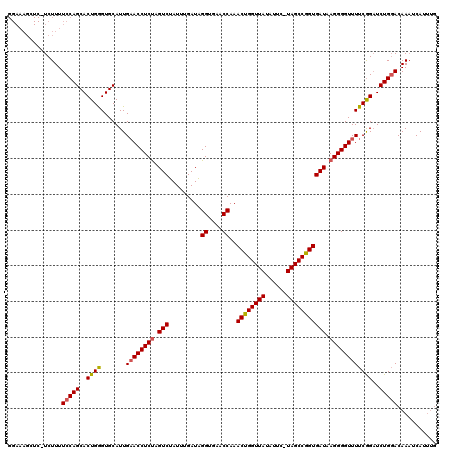
| Location | 15,637,262 – 15,637,381 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.68 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -29.07 |
| Energy contribution | -29.41 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15637262 119 - 22224390 GGAAAGCUCAUCUUUUCCAGCACUGGGUGCAUUGUACCUCCAGUCGAUUUGAUUGGUGAACCAAACUGGUUAUAUUU-UAGCCAGUGAUAAGGGGUUUUCGGAUCUGGACAAAGCAUUUG ((((((......)))))).((...((((((...))))))...((((((((((((((....))))((((((((.....-))))))))............)))))))..)))...))..... ( -35.70) >DroSec_CAF1 1977 115 - 1 GGAAAGCUCGUCUUUUCCAGCACUGAGUGCAUUGAACCUCUAGUCUAUUUGAUAGGUGAACCAAACUGGUUAUAUUC-UAGCCGGUGAUAAGGGGUUUUCAGCUCUGCACAAAUCA---- ((((((......)))))).(((..((((.....((((((((.(((.........((....))..((((((((.....-))))))))))).))))))))...)))))))........---- ( -36.50) >DroSim_CAF1 1980 110 - 1 ----------UCUUUUCCAGCACUGGGUGCAUUGAACCUCUAGUCUAUUUGAUAGGUGAACCAAACUGGUUAUAUUCUUAGCCGGUGAUAAGGGGUUUUCGGAUCUGGACAAUUCAUUUG ----------(((..(((.((((...))))...((((((((.(((.........((....))..((((((((......))))))))))).))))))))..)))...)))........... ( -33.70) >consensus GGAAAGCUC_UCUUUUCCAGCACUGGGUGCAUUGAACCUCUAGUCUAUUUGAUAGGUGAACCAAACUGGUUAUAUUC_UAGCCGGUGAUAAGGGGUUUUCGGAUCUGGACAAAUCAUUUG ...............(((((..((((.......((((((((.(((.........((....))..((((((((......))))))))))).))))))))))))..)))))........... (-29.07 = -29.41 + 0.34)

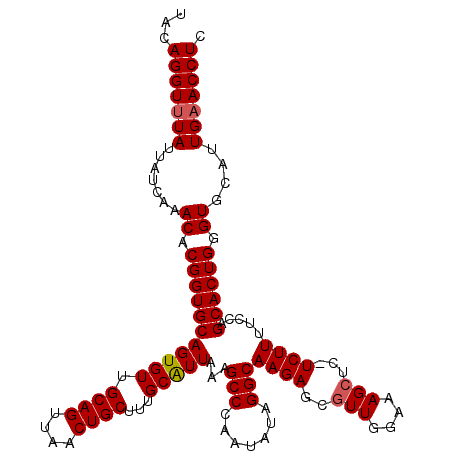
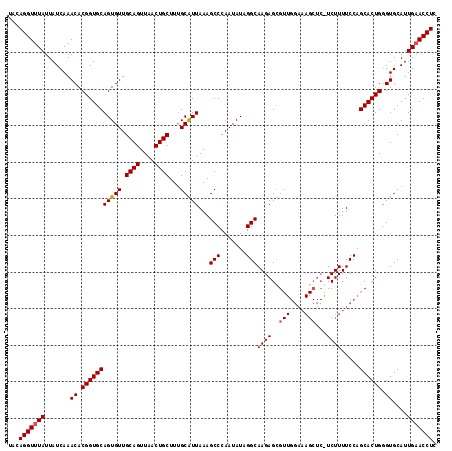
| Location | 15,637,341 – 15,637,458 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -28.12 |
| Energy contribution | -28.73 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15637341 117 - 22224390 UACAGGUUUAUUAUCAAACACGGUGCAGUGUUGCAGUUAACUGCUUAGCAUUAAAGCCCAAUAUAGGCAAGAGUGUUGGAAAGCUCAUCUUUUCCAGCACUGGGUGCAUUGUACCUC .....((((......))))..((((((((...((((....))))...(((((...(((.......)))...((((((((((((......)))))))))))).))))))))))))).. ( -40.30) >DroSec_CAF1 2052 117 - 1 UACAGGUUUAUUAUCAAACACGGUGCAGUGUUGCAGUUAACUGCUUUGCAUUAAAGCCCAAUAUAGGCAAGAGCGUUGGAAAGCUCGUCUUUUCCAGCACUGAGUGCAUUGAACCUC ...(((((((......(((((......)))))((((....))))..((((((...(((.......)))...((.(((((((((......))))))))).)).)))))).))))))). ( -36.60) >DroSim_CAF1 2060 105 - 1 UACAGGUUUAUUAUCAAACACGGUGCAGUGUUGCAGUUAACUGCUUUGCGUUAAAGCCCAAUAUAGGCAAGAGCG------------UCUUUUCCAGCACUGGGUGCAUUGAACCUC ...(((((((.......((.((((((.(((..((((....))))..)))(((...(((.......)))...))).------------.........)))))).))....))))))). ( -30.50) >consensus UACAGGUUUAUUAUCAAACACGGUGCAGUGUUGCAGUUAACUGCUUUGCAUUAAAGCCCAAUAUAGGCAAGAGCGUUGGAAAGCUC_UCUUUUCCAGCACUGGGUGCAUUGAACCUC ...(((((((.......((.(((((((((((.((((....))))...)))))...(((.......)))((((..(((....)))...)))).....)))))).))....))))))). (-28.12 = -28.73 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:01 2006