
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,616,463 – 15,616,733 |
| Length | 270 |
| Max. P | 0.974381 |

| Location | 15,616,463 – 15,616,573 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -16.20 |
| Energy contribution | -18.20 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15616463 110 + 22224390 UAAACCAACCUAUCUCAAACAUCUUGGGAUCUGGGCUACAACAUCCCAAUUUACUUUGCUAAUAUCGUAUUGUAAGUCGUUAUGGUAAAUACCA-GUACCAUCUAUACUUA ...........(((((((.....)))))))((((.......(((..(.((((((..(((.......)))..)))))).)..))).......)))-)............... ( -19.14) >DroSec_CAF1 4649 88 + 1 -------------------CAU----GUAUCUGGGCUCCAACAUCCCAAUUCACUUUGCUAAUAUCGUAUCGUAAGUCGUAAUGGUGAGUACCAGGUACCAUCUAUACUUA -------------------...----((((((((((((...(((..(.(((.((..(((.......)))..)).))).)..)))..)))).))))))))............ ( -20.40) >DroSim_CAF1 4637 107 + 1 UAAA----UCUGGCUUAAACAUCUUGGGAUCUGGGCUCCAACAUCCCAAUUUACUUUGCUAAUAUCGUAUCGUAAGUCGUAAUGGUGAGUACCAGGUACCAUCUAUACUUA ....----................(((.((((((((((...(((..(.((((((..(((.......)))..)))))).)..)))..)))).)))))).))).......... ( -24.60) >consensus UAAA_____CU__CU_AAACAUCUUGGGAUCUGGGCUCCAACAUCCCAAUUUACUUUGCUAAUAUCGUAUCGUAAGUCGUAAUGGUGAGUACCAGGUACCAUCUAUACUUA ........................(((.((((((((((...(((..(.((((((..(((.......)))..)))))).)..)))..)))).)))))).))).......... (-16.20 = -18.20 + 2.00)



| Location | 15,616,573 – 15,616,693 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -38.02 |
| Energy contribution | -38.38 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15616573 120 + 22224390 UUUUUACUUACUCGAGCUAAGCGCCCGCGUUUAAGAAAAACGGUUGGGCCGUCGCGACCUGAGUUGCGAAUGUGCUGCUCCUUGAUUUCGAUGGUAUCGUCGGAGAGAUAGUAGCACAGC ....................((((((((((((.....)))))..)))))..(((((((....))))))).(((((((((.(((...(((((((....))))))))))..))))))))))) ( -40.30) >DroSec_CAF1 4737 120 + 1 UAUUUACUUACUCGCGCUAAGCGCCCGCGUUUGAGAAAAACGGUUGGGCCGUCGCGACCUGAGUUGCGAAUGUGCUGCUCCUUGAUUUCGAUGGUAUCGUCGGAGAGGUAGUAGCACACC .............((.....))((((((((((.....)))))..)))))..(((((((....))))))).(((((((((((((...(((((((....))))))))))).))))))))).. ( -47.30) >DroSim_CAF1 4744 120 + 1 UAUUUACUUACUCGCGCUAAGCGACCGCGUUUGAGAAAAACGGUUGGGCCGUCGCGACCUGAGUUGCGAAUGUGCUGCUCCUUGAUCUCGAUGGUAUCGUCGGAGAGGUAGUAGCACACC ......((((..((((.........))))..)))).....(((.....)))(((((((....))))))).(((((((((((((..((.(((((....))))))))))).))))))))).. ( -44.50) >DroEre_CAF1 4957 114 + 1 ------CUUACCCGAGCUAACCGUCCACGUUUGAGAAAAACGGUUGGGCCGUCGCGUCCCGAGUUGCGAAUGUGUUGCUCCUUGAUUUCGAUGGUAUCAUCGGAGAGGUAGUAACACAGC ------......((..((((((((...(......)....))))))))..))(((((........))))).(((((((((((((...(((((((....))))))))))).))))))))).. ( -38.80) >DroYak_CAF1 4430 114 + 1 ------CAUACCCGAGCCAAGCGUCCACGUUUGAGGAAAACGGUGGGUCCGUCGCGUCCUGAGUUGCGAAUGUGCUGCUCCUUGAUCUCGAUGGUGUCAUCGGAGAGGUAGUAGCACAGC ------......(((((((((((....))))))((((..((((.....))))....))))..))).))..(((((((((((((..((.(((((....))))))))))).))))))))).. ( -42.10) >consensus U_UUUACUUACUCGAGCUAAGCGCCCGCGUUUGAGAAAAACGGUUGGGCCGUCGCGACCUGAGUUGCGAAUGUGCUGCUCCUUGAUUUCGAUGGUAUCGUCGGAGAGGUAGUAGCACAGC ......................((((.(((((.....)))))...))))..(((((((....))))))).(((((((((((((....((((((....)))))).)))).))))))))).. (-38.02 = -38.38 + 0.36)

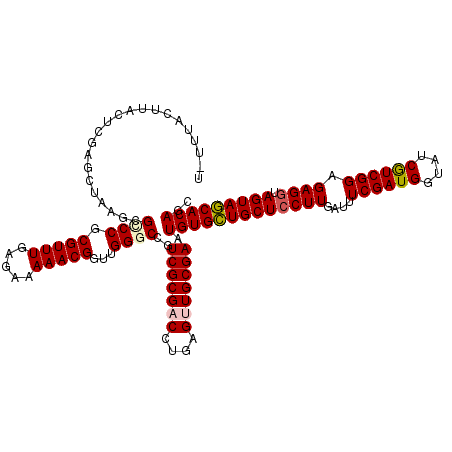
| Location | 15,616,613 – 15,616,733 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -45.85 |
| Consensus MFE | -40.80 |
| Energy contribution | -40.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15616613 120 + 22224390 CGGUUGGGCCGUCGCGACCUGAGUUGCGAAUGUGCUGCUCCUUGAUUUCGAUGGUAUCGUCGGAGAGAUAGUAGCACAGCUCCAGCUUGCGUACGUCUCCGAACUCGGUUCUAUCGUCCC .((((((((..(((((((....))))))).(((((((((.(((...(((((((....))))))))))..))))))))))).)))))).(((.......(((....)))......)))... ( -44.12) >DroSec_CAF1 4777 120 + 1 CGGUUGGGCCGUCGCGACCUGAGUUGCGAAUGUGCUGCUCCUUGAUUUCGAUGGUAUCGUCGGAGAGGUAGUAGCACACCUCCAGCUUGCGUACGUCUCCGAACUCGGUUCUAUCGUCCC .(((((((...(((((((....))))))).(((((((((((((...(((((((....))))))))))).)))))))))..))))))).(((.......(((....)))......)))... ( -46.42) >DroSim_CAF1 4784 120 + 1 CGGUUGGGCCGUCGCGACCUGAGUUGCGAAUGUGCUGCUCCUUGAUCUCGAUGGUAUCGUCGGAGAGGUAGUAGCACACCUCCAGCUUCCGCACGUCUCCAAACUCGGUUCUAUCGUCCC ((((.(((((((((((((....))))))).(((((((((((((..((.(((((....))))))))))).)))))))))......((....)).............)))))).)))).... ( -45.20) >DroEre_CAF1 4991 120 + 1 CGGUUGGGCCGUCGCGUCCCGAGUUGCGAAUGUGUUGCUCCUUGAUUUCGAUGGUAUCAUCGGAGAGGUAGUAACACAGCUCCAGCUUUCGCACAUCUCCGAACUCGGUUCUAUCAUCCC .(((.((((((....((.(.(((.(((((((((((((((((((...(((((((....))))))))))).)))))))))((....)).))))))...))).).)).)))))).)))..... ( -44.90) >DroYak_CAF1 4464 120 + 1 CGGUGGGUCCGUCGCGUCCUGAGUUGCGAAUGUGCUGCUCCUUGAUCUCGAUGGUGUCAUCGGAGAGGUAGUAGCACAGCUCCAGCUUGCGCACAUCUCCGAACUCGGUUCUAUCGUCCC (((((((...((.((((...((((((.((.(((((((((((((..((.(((((....))))))))))).)))))))))..))))))))))))))....(((....))).))))))).... ( -48.60) >consensus CGGUUGGGCCGUCGCGACCUGAGUUGCGAAUGUGCUGCUCCUUGAUUUCGAUGGUAUCGUCGGAGAGGUAGUAGCACAGCUCCAGCUUGCGCACGUCUCCGAACUCGGUUCUAUCGUCCC ((((.((((((..(((....((((((.((.(((((((((((((....((((((....)))))).)))).)))))))))..)))))))).))).............)))))).)))).... (-40.80 = -40.60 + -0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:52 2006