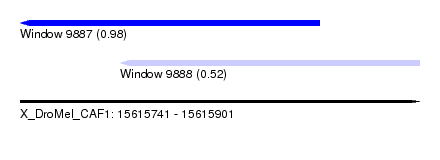
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,615,741 – 15,615,901 |
| Length | 160 |
| Max. P | 0.975812 |

| Location | 15,615,741 – 15,615,861 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -33.32 |
| Energy contribution | -33.04 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15615741 120 - 22224390 CUACUGCGUGGAUCCGCUGAACACUGGAAACAAGGAGAUUCAUUACUAUAGGGAAAAGGAUCUCCAGAUAGGCAGUGUGAUAAACGUGUAUGGCCGUGCGGUGGUGAUAACCGAACUGGA .(((.((((..((((((((.....((....)).((((((((.((.((....)).)).)))))))).......))))).)))..)))))))...((((.((((.......)))).)).)). ( -37.30) >DroSec_CAF1 4142 120 - 1 CUAUUGCGUGGAUCCGCUGAACACCGGAAACAAGGAGAUCCAUUACUAUAGGGAAAAGGAUCUCCAGAUAGGCAGUGUGAUAAACGUGUAUGGCCGUGCGGUGGUGAUAACCGAACUGGA (((((((((..((((((((....(((....)..((((((((.((.((....)).)).)))))))).....))))))).)))..))))).))))((((.((((.......)))).)).)). ( -40.50) >DroSim_CAF1 4126 120 - 1 CUAUUGCGUGGAUCCGCUGAACACCGGAAACAAGGAGAUCCAUUACUAUAGGGAAAAGGAUCUCCAGAUAGGCAGUGUGAUAAACGUGUAUGGCCGUGCGGUGGUGAUAACCGAACUGGA (((((((((..((((((((....(((....)..((((((((.((.((....)).)).)))))))).....))))))).)))..))))).))))((((.((((.......)))).)).)). ( -40.50) >DroEre_CAF1 4241 120 - 1 CUACUGCGUGGAUCCACUGAACACCGGAAACAAGGAUAUCCAUUACUACAGGGAAAAGGAUCUGCAGAUAGGCAGUGUGAUAAACGUUUAUGGUCGUGCGGUGGUGAUAACCGAACUGGA ((((((((..(((((.(((......(....)..((....)).......)))......)))))..)..((((((.((.......))))))))......)))))))......((.....)). ( -29.90) >DroYak_CAF1 4162 120 - 1 CUACUGCGUCGAUCCGUUGAACACCGGAAACAAGGAGAUCCAUUACUAUAAGGAAAAGGAUCUGCAGAUAGGCAGUGUGAUAAACGCUUAUGGUCGUGCGGUGGUGAUAACCGAACUGGA ((((((((.(((((((((..((((.(....)..(.((((((.((.((....)).)).)))))).).........))))....)))).....)))))))))))))......((.....)). ( -34.90) >consensus CUACUGCGUGGAUCCGCUGAACACCGGAAACAAGGAGAUCCAUUACUAUAGGGAAAAGGAUCUCCAGAUAGGCAGUGUGAUAAACGUGUAUGGCCGUGCGGUGGUGAUAACCGAACUGGA .....((((..((((((((......(....)..((((((((.((.((....)).)).)))))))).......))))).)))..))))......((((.((((.......)))).)).)). (-33.32 = -33.04 + -0.28)


| Location | 15,615,781 – 15,615,901 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -30.82 |
| Energy contribution | -31.06 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15615781 120 - 22224390 CGCUACUGAACGUCCUGGGCACCAACAUGCGCAACGUGCGCUACUGCGUGGAUCCGCUGAACACUGGAAACAAGGAGAUUCAUUACUAUAGGGAAAAGGAUCUCCAGAUAGGCAGUGUGA (((.((((...((((((.(((......))).))....((((....))))))))((.((......((....)).((((((((.((.((....)).)).))))))))))...))))))))). ( -34.90) >DroSec_CAF1 4182 120 - 1 CGCUACUGAACGUCCUGGGCACCAAUAUGCGCAACGUGCGCUAUUGCGUGGAUCCGCUGAACACCGGAAACAAGGAGAUCCAUUACUAUAGGGAAAAGGAUCUCCAGAUAGGCAGUGUGA (((.((((...(((((((.((((((((.((((.....))))))))).)))..((....))...))))......((((((((.((.((....)).)).)))))))).)))...))))))). ( -41.00) >DroSim_CAF1 4166 120 - 1 CGCUACUGAACGUCCUGGGCACCAAUAUGCGCAACGUGCGCUAUUGCGUGGAUCCGCUGAACACCGGAAACAAGGAGAUCCAUUACUAUAGGGAAAAGGAUCUCCAGAUAGGCAGUGUGA (((.((((...(((((((.((((((((.((((.....))))))))).)))..((....))...))))......((((((((.((.((....)).)).)))))))).)))...))))))). ( -41.00) >DroEre_CAF1 4281 120 - 1 CGCUACUUAAUGUCCUGGGCACCAACAUGCGCAACGUACGCUACUGCGUGGAUCCACUGAACACCGGAAACAAGGAUAUCCAUUACUACAGGGAAAAGGAUCUGCAGAUAGGCAGUGUGA (((........((((((.(((......))).)....(((((....)))))..(((.(((......(....)..((....)).......))))))..)))))((((......)))).))). ( -30.40) >DroYak_CAF1 4202 120 - 1 CGCUGCUGAAUGUGUUGGGCACCAACAUGCGCAACGUACGCUACUGCGUCGAUCCGUUGAACACCGGAAACAAGGAGAUCCAUUACUAUAAGGAAAAGGAUCUGCAGAUAGGCAGUGUGA (((((((...(((((((.(((......))).))))(.((((....)))))..((((........)))).))).(.((((((.((.((....)).)).)))))).).....)))))))... ( -38.30) >consensus CGCUACUGAACGUCCUGGGCACCAACAUGCGCAACGUGCGCUACUGCGUGGAUCCGCUGAACACCGGAAACAAGGAGAUCCAUUACUAUAGGGAAAAGGAUCUCCAGAUAGGCAGUGUGA .........((((..((.(((......))).)))))).(((.(((((.((..((((........))))..)).((((((((.((.((....)).)).))))))))......)))))))). (-30.82 = -31.06 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:49 2006