
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,614,781 – 15,615,021 |
| Length | 240 |
| Max. P | 0.999712 |

| Location | 15,614,781 – 15,614,901 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -48.78 |
| Consensus MFE | -40.94 |
| Energy contribution | -41.74 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15614781 120 + 22224390 AAGGAGAAUGGCAGGCGACAUCCCCGCAGUGCCGAUCUCACCUUAACUUCGGGCAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCAC ...((((.(((((.(((.......)))..))))).)))).(((...(....)..)))......(((.((.((((..((((.(((((((.....))))))).))))..)))).))..))). ( -49.50) >DroSec_CAF1 3182 120 + 1 AGGGAGAAUGGCAGGCGACAGCCCCGCAGUGCCGAUCUCACCUUAACCUCGGGCAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCAC (((((((.(((((.(((.......)))..))))).)))).))).......((.((....)).))((.((.((((..((((.(((((((.....))))))).))))..)))).))..)).. ( -49.80) >DroSim_CAF1 3166 120 + 1 AGGGAGAAUGGCAAGCGACAGCCCCGCAGUGCCGAUCUCACCUUAACCUCGGGAAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCAC (((((((.(((((.(((.......)))..))))).)))).)))....(((((((.....(((((....)))))...((((.(((((((.....))))))).)))))))))))........ ( -49.30) >DroEre_CAF1 3281 120 + 1 AGGGAGAAUGGGAGACGACAGCCCCGCAGUGCCGACCUCACCUUGGAUUCGGGCAGGAAUGGCUGGUUGGUCGGGUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCAC ..((((....(....)..(((((...(..((((((((.......))..)).)))).)...)))))..((.(((((.((((.(((((((.....))))))).)))).))))).)))))).. ( -49.30) >DroYak_CAF1 3202 120 + 1 AGCGAGAAUGGCAGACGACAGCCCCGCAGUGCCGAUCUCACCUUGGACUCGGGCAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCAC ...((((.(((((..((.......))...))))).))))....((((.((((.((....)).)))).((.((((..((((.(((((((.....))))))).))))..)))).)).)))). ( -46.00) >consensus AGGGAGAAUGGCAGGCGACAGCCCCGCAGUGCCGAUCUCACCUUAACCUCGGGCAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCAC ...((((.(((((.(((.......)))..))))).)))).((........))...((((((.((....))((((..((((.(((((((.....))))))).))))..)))).)))))).. (-40.94 = -41.74 + 0.80)

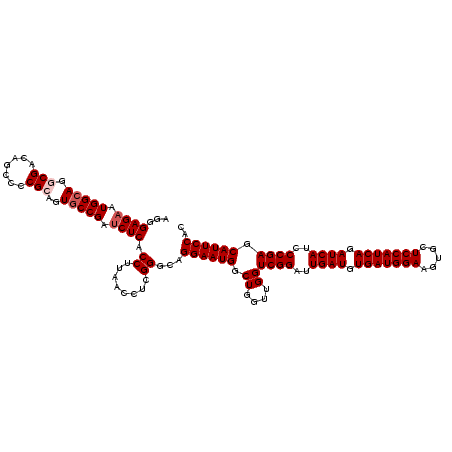

| Location | 15,614,781 – 15,614,901 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -43.42 |
| Consensus MFE | -40.41 |
| Energy contribution | -40.09 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15614781 120 - 22224390 GUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUGCCCGAAGUUAAGGUGAGAUCGGCACUGCGGGGAUGUCGCCUGCCAUUCUCCUU ((((...(((((((.((((.((((((((...)))))))).)))).))))).......(.(((((((((((((..((.....))..))))....))))))))).)))...))))....... ( -41.80) >DroSec_CAF1 3182 120 - 1 GUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUGCCCGAGGUUAAGGUGAGAUCGGCACUGCGGGGCUGUCGCCUGCCAUUCUCCCU ..((((((((((((.((((.((((((((...)))))))).)))).))))).......).))))))............(((.((((..((((..((((.....)))).))))..))))))) ( -43.21) >DroSim_CAF1 3166 120 - 1 GUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUUCCCGAGGUUAAGGUGAGAUCGGCACUGCGGGGCUGUCGCUUGCCAUUCUCCCU ..((((((((((((.((((.((((((((...)))))))).)))).))))).......).))))))............(((.((((..((((..((((.....)))).))))..))))))) ( -44.11) >DroEre_CAF1 3281 120 - 1 GUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAACCCGACCAACCAGCCAUUCCUGCCCGAAUCCAAGGUGAGGUCGGCACUGCGGGGCUGUCGUCUCCCAUUCUCCCU ((((...(((((((.((((.((((((((...)))))))).)))).))))).......(((...(((..((........))..)))..)))...))(((((....)))))))))....... ( -43.00) >DroYak_CAF1 3202 120 - 1 GUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUGCCCGAGUCCAAGGUGAGAUCGGCACUGCGGGGCUGUCGUCUGCCAUUCUCGCU .(((...(((((((.((((.((((((((...)))))))).))))...........(((......))))))))))))).(((((((..((((..((((.....)))).))))..))))))) ( -45.00) >consensus GUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUGCCCGAGGUUAAGGUGAGAUCGGCACUGCGGGGCUGUCGCCUGCCAUUCUCCCU ..((((((((((((.((((.((((((((...)))))))).)))).))))).......).))))))............(((.((((..((((..((((.....)))).))))..))))))) (-40.41 = -40.09 + -0.32)



| Location | 15,614,821 – 15,614,941 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -51.14 |
| Consensus MFE | -46.62 |
| Energy contribution | -46.78 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15614821 120 + 22224390 CCUUAACUUCGGGCAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCACAGGGUUCUCUUAAGCUCGAGAUGAGCCGCCGCCCGCACCG .........(((((.((...((((((.((.((((..((((.(((((((.....))))))).))))..)))).))..)))...(((((((((......)))).)))))))).)).)).))) ( -51.40) >DroSec_CAF1 3222 120 + 1 CCUUAACCUCGGGCAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCACAGGGUUCUCUUCAGCUCGAGAUGAGCCGCCGCCCGCACCG .........(((((.((...((((((.((.((((..((((.(((((((.....))))))).))))..)))).))..)))...(((((((((......)))).)))))))).)).)).))) ( -51.20) >DroSim_CAF1 3206 120 + 1 CCUUAACCUCGGGAAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCACAGGGUUCUCUUCAGCUCGAGAUGAGCCGCCGCCCGCACCG .........(((...((..(((((((.((.((((..((((.(((((((.....))))))).))))..)))).))..)))...(((((((((......)))).))))))))).))...))) ( -48.80) >DroEre_CAF1 3321 120 + 1 CCUUGGAUUCGGGCAGGAAUGGCUGGUUGGUCGGGUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCACAGGGCUUUCUUUAGCUCCAGAUGCGCCGCCGCCCGCACGG ((.((....(((((.((...((((((.((.(((((.((((.(((((((.....))))))).)))).))))).))..)))..(((((......))).))......))).))))))))).)) ( -53.90) >DroYak_CAF1 3242 120 + 1 CCUUGGACUCGGGCAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCACAGGGCUCUCUUCAGCUCCAGAUGUGCAGCCGCCCGCACCG ....((...(((((......(((((..((.((((..((((.(((((((.....))))))).))))..)))).))...(((((((((......)))))....))))))))))))))..)). ( -50.40) >consensus CCUUAACCUCGGGCAGGAAUGGCUGGUUGGUCGGAUUGAUGUGAUGGAAGUGCUCCAUCAGAUCAUCCCGAGCAUUCCACAGGGUUCUCUUCAGCUCGAGAUGAGCCGCCGCCCGCACCG .........(((((.((...((((((.((.((((..((((.(((((((.....))))))).))))..)))).))..)))..(((((......))))).......))).)))))))..... (-46.62 = -46.78 + 0.16)

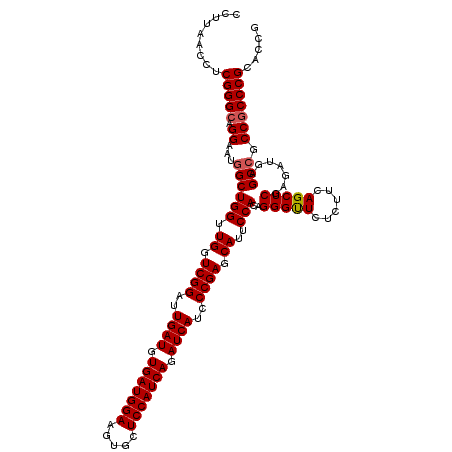

| Location | 15,614,821 – 15,614,941 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -46.88 |
| Consensus MFE | -42.36 |
| Energy contribution | -43.20 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15614821 120 - 22224390 CGGUGCGGGCGGCGGCUCAUCUCGAGCUUAAGAGAACCCUGUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUGCCCGAAGUUAAGG (((.(((((.((((((((.....))))).............(((..((.(((((.((((.((((((((...)))))))).)))).))))).)).))))))...))))))))......... ( -46.90) >DroSec_CAF1 3222 120 - 1 CGGUGCGGGCGGCGGCUCAUCUCGAGCUGAAGAGAACCCUGUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUGCCCGAGGUUAAGG (((.(((((.((((((((.....))))).............(((..((.(((((.((((.((((((((...)))))))).)))).))))).)).))))))...))))))))......... ( -47.50) >DroSim_CAF1 3206 120 - 1 CGGUGCGGGCGGCGGCUCAUCUCGAGCUGAAGAGAACCCUGUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUUCCCGAGGUUAAGG (((.((((((..((((((.....))))))..).....)))))((((((((((((.((((.((((((((...)))))))).)))).))))).......).))))))...)))......... ( -41.71) >DroEre_CAF1 3321 120 - 1 CCGUGCGGGCGGCGGCGCAUCUGGAGCUAAAGAAAGCCCUGUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAACCCGACCAACCAGCCAUUCCUGCCCGAAUCCAAGG .....(((((((.(((((((((((.(((......)))))...)).))))(((((.((((.((((((((...)))))))).)))).))))).......)))....)))))))......... ( -48.50) >DroYak_CAF1 3242 120 - 1 CGGUGCGGGCGGCUGCACAUCUGGAGCUGAAGAGAGCCCUGUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUGCCCGAGUCCAAGG (((.(((((.(((((((((...((.(((......)))))))))...((.(((((.((((.((((((((...)))))))).)))).))))).))..)))))...))))))))......... ( -49.80) >consensus CGGUGCGGGCGGCGGCUCAUCUCGAGCUGAAGAGAACCCUGUGGAAUGCUCGGGAUGAUCUGAUGGAGCACUUCCAUCACAUCAAUCCGACCAACCAGCCAUUCCUGCCCGAGGUUAAGG (((.(((((.((((((((.....))))).............(((..((.(((((.((((.((((((((...)))))))).)))).))))).)).))))))...))))))))......... (-42.36 = -43.20 + 0.84)

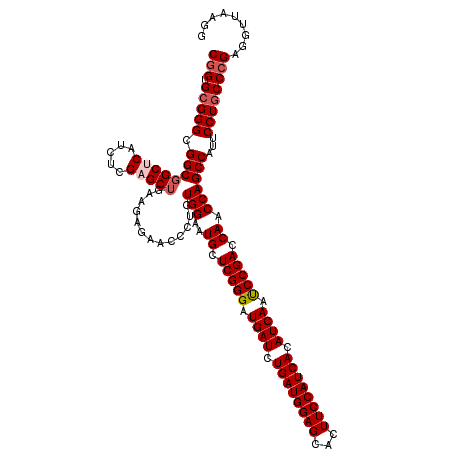
| Location | 15,614,901 – 15,615,021 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -48.32 |
| Consensus MFE | -38.80 |
| Energy contribution | -38.92 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15614901 120 - 22224390 GAUCAGCGAUCACGAGAUCAUCAGCCUCUGCCGGCACUUUUCGGCGGAGAAGAGUCAACCAAAUGCCUGCGAUCGGUCCACGGUGCGGGCGGCGGCUCAUCUCGAGCUUAAGAGAACCCU ((((...)))).((((((.......(((((((((......)))))))))..(((((..((....((((((.((((.....)))))))))))).)))))))))))................ ( -45.80) >DroSec_CAF1 3302 120 - 1 GAUCAGCGAUCACGAGAUCAUUAGCCUCUGCCGGCACUUUUCGGCGGAGAAGAGUCAACCAAAUGCCUGCGAUCGAUCCACGGUGCGGGCGGCGGCUCAUCUCGAGCUGAAGAGAACCCU ..(((((.....((((((.......(((((((((......)))))))))..(((((..((....((((((.((((.....)))))))))))).))))))))))).))))).......... ( -48.70) >DroSim_CAF1 3286 120 - 1 GAUCAGUGAUCACGAGAUCAUCAGCCUCUGCCGGCACUUUUCGGCGGAGAAGAGUCAGCCAAAUGCCUGCGAUCGAUCCACGGUGCGGGCGGCGGCUCAUCUCGAGCUGAAGAGAACCCU .....((((((....))))))..(((......))).(((((((((.((((.(((((.(((....((((((.((((.....)))))))))))))))))).))))..)))))))))...... ( -53.50) >DroEre_CAF1 3401 120 - 1 GAUCAGUGAUCAUGAGAUCAUCAGUCUCUGCCGCCACUUUUCGGCGGAGAAGAGUCAACCAAAUGCCUGCGACCGAUCCACCGUGCGGGCGGCGGCGCAUCUGGAGCUAAAGAAAGCCCU (((..((((((....))))))..)))...(((((((((((((......))))))).........(((((((..((......)))))))))))))))......((.(((......))))). ( -46.00) >DroYak_CAF1 3322 120 - 1 GAUCAGCGAUCACGAGAUCAUCAGCCUCUGUCGGCACUUCUCUGCGGAGAAGAGUCAACCGAAUGCCUGCGAUCGAUCCACGGUGCGGGCGGCUGCACAUCUGGAGCUGAAGAGAGCCCU ..(((((((((....))))..(((((((.((.(((.((((((....)))))).))).)).))..((((((.((((.....)))))))))))))))..........))))).......... ( -47.60) >consensus GAUCAGCGAUCACGAGAUCAUCAGCCUCUGCCGGCACUUUUCGGCGGAGAAGAGUCAACCAAAUGCCUGCGAUCGAUCCACGGUGCGGGCGGCGGCUCAUCUCGAGCUGAAGAGAACCCU ..(((((((((....))))....(((...((((((.((((((....)))))).)))........((((((.((((.....)))))))))))))))).........))))).......... (-38.80 = -38.92 + 0.12)

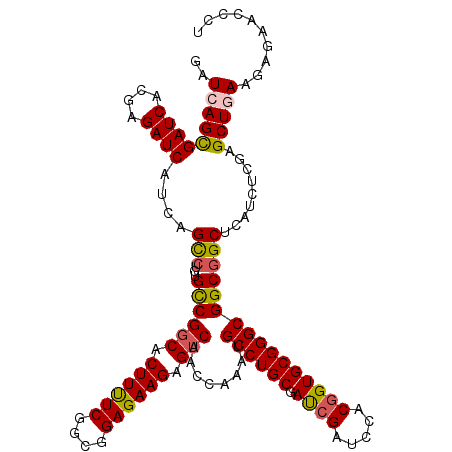

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:45 2006